Structural Basis for Multi-specificity of MRG Domains
- PMID: 25960410
- PMCID: PMC4456287
- DOI: 10.1016/j.str.2015.03.020
Structural Basis for Multi-specificity of MRG Domains
Abstract
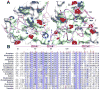
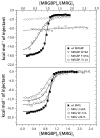
Chromatin-binding proteins play vital roles in the assembly and recruitment of multi-subunit complexes harboring effector proteins to specific genomic loci. MRG15, a chromodomain-containing chromatin-binding protein, recruits diverse chromatin-associated complexes that regulate gene transcription, DNA repair, and RNA splicing. Previous studies with Pf1, another chromatin-binding subunit of the Sin3S/Rpd3S histone deacetylase complex, defined the sequence and structural requirements for interactions with the MRG15 MRG domain, a common target of diverse subunits in the aforementioned complexes. We now show that MRGBP, a member of the Tip60/NuA4 histone acetyltransferase complex, engages the same two surfaces of the MRG domain as Pf1. High-affinity interactions occur via a bipartite structural motif including an FxLP sequence motif. MRGBP shares little sequence and structural similarity with Pf1, yet targets similar pockets on the surface of the MRG domain, mimicking Pf1 in its interactions. Our studies shed light onto how MRG domains have evolved to bind diverse targets.
Copyright © 2015 Elsevier Ltd. All rights reserved.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Bax A, Grzesiek S. Methodological advances in protein NMR. Accounts Chem Res. 1993;26:131–138.
-
- Bowman BR, Moure CM, Kirtane BM, Welschhans RL, Tominaga K, Pereira-Smith OM, Quiocho FA. Multipurpose MRG domain involved in cell senescence and proliferation exhibits structural homology to a DNA-interacting domain. Structure. 2006;14:151–158. - PubMed
-
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. - PubMed
-
- Cai Y, Jin J, Tomomori-Sato C, Sato S, Sorokina I, Parmely TJ, Conaway RC, Conaway JW. Identification of new subunits of the multiprotein mammalian TRRAP/TIP60-containing histone acetyltransferase complex. J Biol Chem. 2003;278:42733–42736. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

