A Practical and Scalable Tool to Find Overlaps between Sequences
- PMID: 25961045
- PMCID: PMC4417569
- DOI: 10.1155/2015/905261
A Practical and Scalable Tool to Find Overlaps between Sequences
Abstract
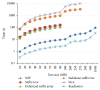
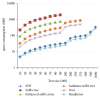
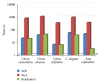
The evolution of the next generation sequencing technology increases the demand for efficient solutions, in terms of space and time, for several bioinformatics problems. This paper presents a practical and easy-to-implement solution for one of these problems, namely, the all-pairs suffix-prefix problem, using a compact prefix tree. The paper demonstrates an efficient construction of this time-efficient and space-economical tree data structure. The paper presents techniques for parallel implementations of the proposed solution. Experimental evaluation indicates superior results in terms of space and time over existing solutions. Results also show that the proposed technique is highly scalable in a parallel execution environment.
Figures

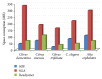
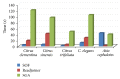




References
-
- Gusfield D., Landau G. M., Schieber B. An efficient algorithm for the all pairs suffix-prefix problem. Information Processing Letters. 1992;41(4):181–185. doi: 10.1016/0020-0190(92)90176-V. - DOI
-
- Gusfield D. Algorithms on Strings, Trees, and Sequences—Computer Science and Computational Biology. Cambridge University Press; 1997.
-
- Weiner P. Linear pattern matching algorithms. Proceedings of the IEEE Conference Record of 14th Annual Symposium on Switching and Automata Theory (SWAT ’08); October 1973; pp. 1–11. - DOI
-
- Manber U., Myers G. Suffix arrays: a new method for on-line string searches. Proceedings of the 1st Annual ACM-SIAM Symposium on Discrete Algorithms (SODA '90); 1990; Philadelphia, Pa, USA. Society for Industrial and Applied Mathematics; pp. 319–327.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

