Uptake of CCR7 by KIR2DS4⁺ NK cells is induced upon recognition of certain HLA-C alleles
- PMID: 25961063
- PMCID: PMC4415677
- DOI: 10.1155/2015/754373
Uptake of CCR7 by KIR2DS4⁺ NK cells is induced upon recognition of certain HLA-C alleles
Abstract
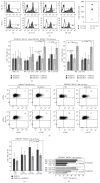
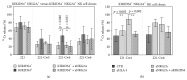
The KIR2DS4 receptor is the oldest KIR2DS expressed by human NK lymphocytes. The specificity of recognition of this receptor for various HLA class I alleles has been demonstrated; however it remains poorly understood whether these interactions may result in the activation of some specific functions in NK cells. Here, we examined the functional outcome of the KIR2DS4/HLA class I interaction by the use of an alternative functional system based on the ability of KIR2DS4 to regulate the mechanism of trogocytosis by NK cells. We demonstrate that KIR2DS4 can induce the uptake of CCR7 by KIR2DS4(+) NKG2A(+) NK cell clones after interacting with CCR7(+) target cells expressing HLA-Cw4 and HLA-Cw6 alleles. However this interaction is not always sufficient to override the inhibition generated by NKG2A expressed on the same NK cells. The recognition of HLA-Cw4 was confirmed by experiments of cytotoxicity against HLA-C-transfected cells. We also show that, different from resting NK cells, the acquisition of CCR7 in response to IL-18 cannot occur in IL2-activated NK cells because of a marked downregulation in their IL-18Rα expression. As a consequence trogocytosis represents the major mechanism by which KIR2DS4(+) activated NK cells acquire the expression of this chemokine receptor.
Figures

Similar articles
-
Recognition of HLA-Cw4 but not HLA-Cw6 by the NK cell receptor killer cell Ig-like receptor two-domain short tail number 4.J Immunol. 2001 Jun 15;166(12):7260-7. doi: 10.4049/jimmunol.166.12.7260. J Immunol. 2001. PMID: 11390475
-
Human NK cell receptor KIR2DS4 detects a conserved bacterial epitope presented by HLA-C.Proc Natl Acad Sci U S A. 2019 Jun 25;116(26):12964-12973. doi: 10.1073/pnas.1903781116. Epub 2019 May 28. Proc Natl Acad Sci U S A. 2019. PMID: 31138701 Free PMC article.
-
Uptake of CCR7 and acquisition of migratory properties by human KIR+ NK cells interacting with monocyte-derived DC or EBV cell lines: regulation by KIR/HLA-class I interaction.Blood. 2009 Nov 5;114(19):4108-16. doi: 10.1182/blood-2009-05-222265. Epub 2009 Sep 11. Blood. 2009. PMID: 19749090
-
Bridging innate NK cell functions with adaptive immunity.Adv Exp Med Biol. 2011;780:45-55. doi: 10.1007/978-1-4419-5632-3_5. Adv Exp Med Biol. 2011. PMID: 21842364 Review.
-
Natural killer cell recognition of HLA class I molecules.Rev Immunogenet. 2000;2(3):433-48. Rev Immunogenet. 2000. PMID: 11256749 Review.
Cited by
-
Activating KIR2DS4 Is Expressed by Uterine NK Cells and Contributes to Successful Pregnancy.J Immunol. 2016 Dec 1;197(11):4292-4300. doi: 10.4049/jimmunol.1601279. Epub 2016 Nov 4. J Immunol. 2016. PMID: 27815424 Free PMC article.
-
Human NK Cell Subsets Redistribution in Pathological Conditions: A Role for CCR7 Receptor.Front Immunol. 2016 Oct 7;7:414. doi: 10.3389/fimmu.2016.00414. eCollection 2016. Front Immunol. 2016. PMID: 27774094 Free PMC article. Review.
-
Cancer Immunotherapy by Blocking Immune Checkpoints on Innate Lymphocytes.Cancers (Basel). 2020 Nov 25;12(12):3504. doi: 10.3390/cancers12123504. Cancers (Basel). 2020. PMID: 33255582 Free PMC article. Review.
-
Features of Memory-Like and PD-1(+) Human NK Cell Subsets.Front Immunol. 2016 Sep 14;7:351. doi: 10.3389/fimmu.2016.00351. eCollection 2016. Front Immunol. 2016. PMID: 27683578 Free PMC article. Review.
-
The early expansion of anergic NKG2Apos/CD56dim/CD16neg natural killer represents a therapeutic target in haploidentical hematopoietic stem cell transplantation.Haematologica. 2018 Aug;103(8):1390-1402. doi: 10.3324/haematol.2017.186619. Epub 2018 Apr 26. Haematologica. 2018. PMID: 29700172 Free PMC article.
References
-
- Sivori S., Vitale M., Bottino C., et al. CD94 functions as a natural killer cell inhibitory receptor for different HLA class I alleles: identification of the inhibitory form of CD94 by the use of novel monoclonal antibodies. European Journal of Immunology. 1996;26(10):2487–2492. doi: 10.1002/eji.1830261032. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

