Discovery of an inhibitor of Z-alpha1 antitrypsin polymerization
- PMID: 25961288
- PMCID: PMC4427445
- DOI: 10.1371/journal.pone.0126256
Discovery of an inhibitor of Z-alpha1 antitrypsin polymerization
Abstract
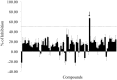
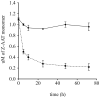
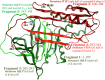
Polymerization of the Z variant alpha-1-antitrypsin (Z-α1AT) results in the most common and severe form of α1AT deficiency (α1ATD), a debilitating genetic disorder whose clinical manifestations range from asymptomatic to fatal liver and/or lung disease. As the altered conformation of Z-α1AT and its attendant aggregation are responsible for pathogenesis, the polymerization process per se has become a major target for the development of therapeutics. Based on the ability of Z-α1AT to aggregate by recruiting the reactive center loop (RCL) of another Z-α1AT into its s4A cavity, we developed a high-throughput screening assay that uses a modified 6-mer peptide mimicking the RCL to screen for inhibitors of Z-α1AT polymer growth. A subset of compounds from the Library of Pharmacologically Active Compounds (LOPAC) with molecular weights ranging from 300 to 700 Da, was used to evaluate the assay's capabilities. The inhibitor S-(4-nitrobenzyl)-6-thioguanosine was identified as a lead compound and its ability to prevent Z-α1AT polymerization confirmed by secondary assays. To further investigate the binding location of S-(4-nitrobenzyl)-6-thioguanosine, an in silico strategy was pursued and the intermediate α1AT M* state modeled to allow molecular docking simulations and explore various potential binding sites. Docking results predict that S-(4-nitrobenzyl)-6-thioguanosine can bind at the s4A cavity and at the edge of β-sheet A. The former binding site would directly block RCL insertion whereas the latter site would prevent β-sheet A from expanding between s3A/s5A, and thus indirectly impede RCL insertion. Altogether, our investigations have revealed a novel compound that inhibits the formation of Z-α1AT polymers, as well as in vitro and in silico strategies for identifying and characterizing additional blocking molecules of Z-α1AT polymerization.
Conflict of interest statement
Figures




References
-
- Brantly M, Nukiwa T, Crystal RG. Molecular basis of alpha-1-antitrypsin deficiency. Am J Med. 1988;84: 13–31. Available: http://www.ncbi.nlm.nih.gov/pubmed/3289385 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

