Condensin confers the longitudinal rigidity of chromosomes
- PMID: 25961503
- PMCID: PMC5207317
- DOI: 10.1038/ncb3167
Condensin confers the longitudinal rigidity of chromosomes
Abstract
In addition to inter-chromatid cohesion, mitotic and meiotic chromatids must have three physical properties: compaction into 'threads' roughly co-linear with their DNA sequence, intra-chromatid cohesion determining their rigidity, and a mechanism to promote sister chromatid disentanglement. A fundamental issue in chromosome biology is whether a single molecular process accounts for all three features. There is universal agreement that a pair of Smc-kleisin complexes called condensin I and II facilitate sister chromatid disentanglement, but whether they also confer thread formation or longitudinal rigidity is either controversial or has never been directly addressed respectively. We show here that condensin II (beta-kleisin) has an essential role in all three processes during meiosis I in mouse oocytes and that its function overlaps with that of condensin I (gamma-kleisin), which is otherwise redundant. Pre-assembled meiotic bivalents unravel when condensin is inactivated by TEV cleavage, proving that it actually holds chromatin fibres together.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
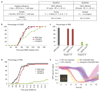
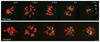

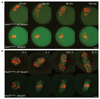

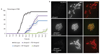
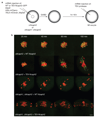
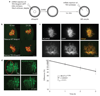
Comment in
-
Clarifying the role of condensin in shaping chromosomes.Nat Cell Biol. 2015 Jun;17(6):711-3. doi: 10.1038/ncb3183. Nat Cell Biol. 2015. PMID: 26022918
-
Chromosome condensation: weaving an untangled web.Curr Biol. 2015 Aug 3;25(15):R663-6. doi: 10.1016/j.cub.2015.06.026. Curr Biol. 2015. PMID: 26241143
References
-
- Cremer T, Tesin D, Hopman AH, Manuelidis L. Rapid interphase and metaphase assessment of specific chromosomal changes in neuroectodermal tumor cells by in situ hybridization with chemically modified DNA probes. Exp Cell Res. 1988;176:199–220. - PubMed
-
- Flemming W. Beitrage zur Kenntnisseder Zelle undIhreer Lebenserscheinungen. Archiv f mikrosk Anatomie. 1880;18:151–259.
-
- Nasmyth K. Segregating sister genomes: the molecular biology of chromosome separation. Science. 2002;297:559–565. - PubMed
-
- Nasmyth K. Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu Rev Genet. 2001;35:673–745. - PubMed
-
- Schleiffer A, et al. Kleisins: a superfamily of bacterial and eukaryotic SMC protein partners. Mol Cell. 2003;11:571–575. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

