Phylogeographical analysis of the dominant multidrug-resistant H58 clade of Salmonella Typhi identifies inter- and intracontinental transmission events
- PMID: 25961941
- PMCID: PMC4921243
- DOI: 10.1038/ng.3281
Phylogeographical analysis of the dominant multidrug-resistant H58 clade of Salmonella Typhi identifies inter- and intracontinental transmission events
Abstract
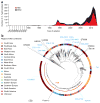
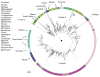
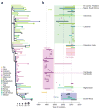
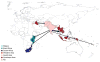
The emergence of multidrug-resistant (MDR) typhoid is a major global health threat affecting many countries where the disease is endemic. Here whole-genome sequence analysis of 1,832 Salmonella enterica serovar Typhi (S. Typhi) identifies a single dominant MDR lineage, H58, that has emerged and spread throughout Asia and Africa over the last 30 years. Our analysis identifies numerous transmissions of H58, including multiple transfers from Asia to Africa and an ongoing, unrecognized MDR epidemic within Africa itself. Notably, our analysis indicates that H58 lineages are displacing antibiotic-sensitive isolates, transforming the global population structure of this pathogen. H58 isolates can harbor a complex MDR element residing either on transmissible IncHI1 plasmids or within multiple chromosomal integration sites. We also identify new mutations that define the H58 lineage. This phylogeographical analysis provides a framework to facilitate global management of MDR typhoid and is applicable to similar MDR lineages emerging in other bacterial species.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


References
-
- Parry CM, Hien TT, Dougan G, White NJ, Farrar JJ. Typhoid fever. N Engl J Med. 2002;347:1770–1782. - PubMed
-
- Connor BA, Schwartz E. Typhoid and paratyphoid fever in travellers. Lancet Infect Dis. 2005;5:623–628. - PubMed
-
- Mogasale V, et al. Burden of typhoid fever in low-income and middle-income countries: a systematic, literature-based update with risk-factor adjustment. Lancet Glob Health. 2014;2:e570–e580. - PubMed
-
- Bodhidatta L, Taylor DN, Thisyakorn U, Echeverria P. Control of typhoid fever in Bangkok, Thailand, by annual immunization of schoolchildren with parenteral typhoid vaccine. Rev Infect Dis. 1987;9:841–845. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 TW009237/TW/FIC NIH HHS/United States
- BB/L018845/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- WT092152MA/WT_/Wellcome Trust/United Kingdom
- MR/J003999/1/MRC_/Medical Research Council/United Kingdom
- R01 AI099525-02/AI/NIAID NIH HHS/United States
- U01 AI062563/AI/NIAID NIH HHS/United States
- R01 AI099525/AI/NIAID NIH HHS/United States
- 089276/WT_/Wellcome Trust/United Kingdom
- BB/L018845/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 101113/Z/13/Z/WT_/Wellcome Trust/United Kingdom
- 001/WHO_/World Health Organization/International
- 100087/WT_/Wellcome Trust/United Kingdom
- G1100100/MRC_/Medical Research Council/United Kingdom
- 089275/H/09/Z/WT_/Wellcome Trust/United Kingdom
- G1100100/1/MRC_/Medical Research Council/United Kingdom
- G9818340/MRC_/Medical Research Council/United Kingdom
- BB/L017679/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 100891/WT_/Wellcome Trust/United Kingdom
- BB/L018926/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/L018926/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 098051/WT_/Wellcome Trust/United Kingdom
- 100087/Z/12/Z/WT_/Wellcome Trust/United Kingdom
- R01 AI097493/AI/NIAID NIH HHS/United States
- BB/J010367/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources

