The impact of low-frequency and rare variants on lipid levels
- PMID: 25961943
- PMCID: PMC4757735
- DOI: 10.1038/ng.3300
The impact of low-frequency and rare variants on lipid levels
Abstract
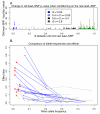
Using a genome-wide screen of 9.6 million genetic variants achieved through 1000 Genomes Project imputation in 62,166 samples, we identify association to lipid traits in 93 loci, including 79 previously identified loci with new lead SNPs and 10 new loci, 15 loci with a low-frequency lead SNP and 10 loci with a missense lead SNP, and 2 loci with an accumulation of rare variants. In six loci, SNPs with established function in lipid genetics (CELSR2, GCKR, LIPC and APOE) or candidate missense mutations with predicted damaging function (CD300LG and TM6SF2) explained the locus associations. The low-frequency variants increased the proportion of variance explained, particularly for low-density lipoprotein cholesterol and total cholesterol. Altogether, our results highlight the impact of low-frequency variants in complex traits and show that imputation offers a cost-effective alternative to resequencing.
Figures



References
-
- Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 2007;38:906–913. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

