Excess of rare, inherited truncating mutations in autism
- PMID: 25961944
- PMCID: PMC4449286
- DOI: 10.1038/ng.3303
Excess of rare, inherited truncating mutations in autism
Abstract
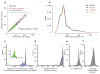
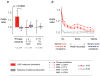
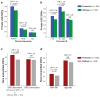
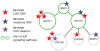
To assess the relative impact of inherited and de novo variants on autism risk, we generated a comprehensive set of exonic single-nucleotide variants (SNVs) and copy number variants (CNVs) from 2,377 families with autism. We find that private, inherited truncating SNVs in conserved genes are enriched in probands (odds ratio = 1.14, P = 0.0002) in comparison to unaffected siblings, an effect involving significant maternal transmission bias to sons. We also observe a bias for inherited CNVs, specifically for small (<100 kb), maternally inherited events (P = 0.01) that are enriched in CHD8 target genes (P = 7.4 × 10(-3)). Using a logistic regression model, we show that private truncating SNVs and rare, inherited CNVs are statistically independent risk factors for autism, with odds ratios of 1.11 (P = 0.0002) and 1.23 (P = 0.01), respectively. This analysis identifies a second class of candidate genes (for example, RIMS1, CUL7 and LZTR1) where transmitted mutations may create a sensitized background but are unlikely to be completely penetrant.
Conflict of interest statement
E.E.E. is on the scientific advisory board (SAB) of DNAnexus, Inc. and is a consultant for Kunming University of Science and Technology (KUST) as part of the 1000 China Talent Program.
Figures

References
-
- Autism, I. MMWR Surveill Summ. 2012:1–9.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

