Small RNA Profiling of Two Important Cultivars of Banana and Overexpression of miRNA156 in Transgenic Banana Plants
- PMID: 25962076
- PMCID: PMC4427177
- DOI: 10.1371/journal.pone.0127179
Small RNA Profiling of Two Important Cultivars of Banana and Overexpression of miRNA156 in Transgenic Banana Plants
Abstract
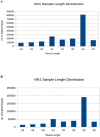
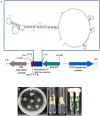
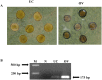
Micro RNAs (miRNAs) are a class of non-coding, short RNAs having important roles in regulation of gene expression. Although plant miRNAs have been studied in detail in some model plants, less is known about these miRNAs in important fruit plants like banana. miRNAs have pivotal roles in plant growth and development, and in responses to diverse biotic and abiotic stress stimuli. Here, we have analyzed the small RNA expression profiles of two different economically significant banana cultivars by using high-throughput sequencing technology. We identified a total of 170 and 244 miRNAs in the two libraries respectively derived from cv. Grand Naine and cv. Rasthali leaves. In addition, several cultivar specific microRNAs along with their putative target transcripts were also detected in our studies. To validate our findings regarding the small RNA profiles, we also undertook overexpression of a common microRNA, MusamiRNA156 in transgenic banana plants. The transgenic plants overexpressing the stem-loop sequence derived from MusamiRNA156 gene were stunted in their growth together with peculiar changes in leaf anatomy. These results provide a foundation for further investigations into important physiological and metabolic pathways operational in banana in general and cultivar specific traits in particular.
Conflict of interest statement
Figures

Similar articles
-
Transcripts and MicroRNAs Responding to Salt Stress in Musa acuminata Colla (AAA Group) cv. Berangan Roots.PLoS One. 2015 May 20;10(5):e0127526. doi: 10.1371/journal.pone.0127526. eCollection 2015. PLoS One. 2015. PMID: 25993649 Free PMC article.
-
Banana sRNAome and degradome identify microRNAs functioning in differential responses to temperature stress.BMC Genomics. 2019 Jan 10;20(1):33. doi: 10.1186/s12864-018-5395-1. BMC Genomics. 2019. PMID: 30630418 Free PMC article.
-
Identification of miRNAs involved in fruit ripening in Cavendish bananas by deep sequencing.BMC Genomics. 2015 Oct 13;16:776. doi: 10.1186/s12864-015-1995-1. BMC Genomics. 2015. PMID: 26462563 Free PMC article.
-
Exploring diverse roles of micro RNAs in banana: Current status and future prospective.Physiol Plant. 2021 Dec;173(4):1323-1334. doi: 10.1111/ppl.13311. Epub 2020 Dec 22. Physiol Plant. 2021. PMID: 33305854 Review.
-
Exploration of small non coding RNAs in wheat (Triticum aestivum L.).Plant Mol Biol. 2012 Sep;80(1):67-73. doi: 10.1007/s11103-011-9835-4. Epub 2011 Oct 19. Plant Mol Biol. 2012. PMID: 22009635 Review.
Cited by
-
Characterization of microRNAs and Target Genes in Musa acuminata subsp. burmannicoides, var. Calcutta 4 during Interaction with Pseudocercospora musae.Plants (Basel). 2023 Mar 28;12(7):1473. doi: 10.3390/plants12071473. Plants (Basel). 2023. PMID: 37050099 Free PMC article.
-
Small RNA and Transcriptome Sequencing Reveal a Potential miRNA-Mediated Interaction Network That Functions during Somatic Embryogenesis in Lilium pumilum DC. Fisch.Front Plant Sci. 2017 Apr 20;8:566. doi: 10.3389/fpls.2017.00566. eCollection 2017. Front Plant Sci. 2017. PMID: 28473835 Free PMC article.
-
Overexpression of native Musa-miR397 enhances plant biomass without compromising abiotic stress tolerance in banana.Sci Rep. 2019 Nov 11;9(1):16434. doi: 10.1038/s41598-019-52858-3. Sci Rep. 2019. PMID: 31712582 Free PMC article.
-
Identification of Fusarium oxysporum f. sp. cubense tropical race 4 (Foc TR4) responsive miRNAs in banana root.Sci Rep. 2019 Sep 23;9(1):13682. doi: 10.1038/s41598-019-50130-2. Sci Rep. 2019. PMID: 31548557 Free PMC article.
-
Computational identification and comparative analysis of miRNA precursors in three palm species.Planta. 2016 May;243(5):1265-77. doi: 10.1007/s00425-016-2486-6. Epub 2016 Feb 26. Planta. 2016. PMID: 26919984
References
-
- Vaucheret H (2006) Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes Dev 20: 759–771. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

