Single-molecule super-resolution imaging of chromosomes and in situ haplotype visualization using Oligopaint FISH probes
- PMID: 25962338
- PMCID: PMC4430122
- DOI: 10.1038/ncomms8147
Single-molecule super-resolution imaging of chromosomes and in situ haplotype visualization using Oligopaint FISH probes
Abstract
Fluorescence in situ hybridization (FISH) is a powerful single-cell technique for studying nuclear structure and organization. Here we report two advances in FISH-based imaging. We first describe the in situ visualization of single-copy regions of the genome using two single-molecule super-resolution methodologies. We then introduce a robust and reliable system that harnesses single-nucleotide polymorphisms (SNPs) to visually distinguish the maternal and paternal homologous chromosomes in mammalian and insect systems. Both of these new technologies are enabled by renewable, bioinformatically designed, oligonucleotide-based Oligopaint probes, which we augment with a strategy that uses secondary oligonucleotides (oligos) to produce and enhance fluorescent signals. These advances should substantially expand the capability to query parent-of-origin-specific chromosome positioning and gene expression on a cell-by-cell basis.
Figures
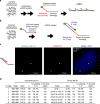
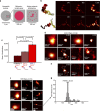

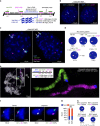
References
-
- van der Ploeg M. Cytochemical nucleic acid research during the twentieth century. Eur. J. Histochem. 44, 7–42 (2000). - PubMed
-
- Levsky J. M. & Singer R. H. Fluorescence in situ hybridization: past, present and future. J. Cell Sci. 116, 2833–2888 (2003). - PubMed
-
- Yamada N. A. et al. Visualization of fine-scale genomic structure by oligonucleotide-based high-resolution FISH. Cytogenet. Genome Res. 132, 248–254 (2011). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM061936/GM/NIGMS NIH HHS/United States
- DP2 OD004641/OD/NIH HHS/United States
- F32CA157188/CA/NCI NIH HHS/United States
- DP1 GM106412/GM/NIGMS NIH HHS/United States
- R01GM61936/GM/NIGMS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- 5R21HD072481/HD/NICHD NIH HHS/United States
- DP2 OD007292/OD/NIH HHS/United States
- 5DP1GM106412/DP/NCCDPHP CDC HHS/United States
- R21 HD072481/HD/NICHD NIH HHS/United States
- 1DP2OD004641/OD/NIH HHS/United States
- R01 EB018659/EB/NIBIB NIH HHS/United States
- 1DP2OD007292/OD/NIH HHS/United States
- R01-GM090278/GM/NIGMS NIH HHS/United States
- F32 CA157188/CA/NCI NIH HHS/United States
- 1R01EB018659/EB/NIBIB NIH HHS/United States
- R01 GM090278/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

