Induction of the Nitrate Assimilation nirA Operon and Protein-Protein Interactions in the Maturation of Nitrate and Nitrite Reductases in the Cyanobacterium Anabaena sp. Strain PCC 7120
- PMID: 25962912
- PMCID: PMC4524197
- DOI: 10.1128/JB.00198-15
Induction of the Nitrate Assimilation nirA Operon and Protein-Protein Interactions in the Maturation of Nitrate and Nitrite Reductases in the Cyanobacterium Anabaena sp. Strain PCC 7120
Abstract
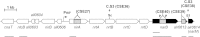
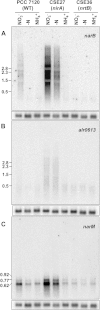
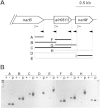
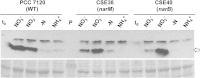
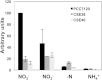
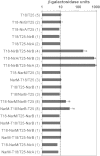
Nitrate is widely used as a nitrogen source by cyanobacteria, in which the nitrate assimilation structural genes frequently constitute the so-called nirA operon. This operon contains the genes encoding nitrite reductase (nirA), a nitrate/nitrite transporter (frequently an ABC-type transporter; nrtABCD), and nitrate reductase (narB). In the model filamentous cyanobacterium Anabaena sp. strain PCC 7120, which can fix N2 in specialized cells termed heterocysts, the nirA operon is expressed at high levels only in media containing nitrate or nitrite and lacking ammonium, a preferred nitrogen source. Here we examined the genes downstream of the nirA operon in Anabaena and found that a small open reading frame of unknown function, alr0613, can be cotranscribed with the operon. The next gene in the genome, alr0614 (narM), showed an expression pattern similar to that of the nirA operon, implying correlated expression of narM and the operon. A mutant of narM with an insertion mutation failed to produce nitrate reductase activity, consistent with the idea that NarM is required for the maturation of NarB. Both narM and narB mutants were impaired in the nitrate-dependent induction of the nirA operon, suggesting that nitrite is an inducer of the operon in Anabaena. It has previously been shown that the nitrite reductase protein NirA requires NirB, a protein likely involved in protein-protein interactions, to attain maximum activity. Bacterial two-hybrid analysis confirmed possible NirA-NirB and NarB-NarM interactions, suggesting that the development of both nitrite reductase and nitrate reductase activities in cyanobacteria involves physical interaction of the corresponding enzymes with their cognate partners, NirB and NarM, respectively.
Importance: Nitrate is an important source of nitrogen for many microorganisms that is utilized through the nitrate assimilation system, which includes nitrate/nitrite membrane transporters and the nitrate and nitrite reductases. Many cyanobacteria assimilate nitrate, but regulation of the nitrate assimilation system varies in different cyanobacterial groups. In the N2-fixing, heterocyst-forming cyanobacteria, the nirA operon, which includes the structural genes for the nitrate assimilation system, is expressed in the presence of nitrate or nitrite if ammonium is not available to the cells. Here we studied the genes required for production of an active nitrate reductase, providing information on the nitrate-dependent induction of the operon, and found evidence for possible protein-protein interactions in the maturation of nitrate reductase and nitrite reductase.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures
Similar articles
-
Negative regulation of expression of the nitrate assimilation nirA operon in the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120.J Bacteriol. 2010 Jun;192(11):2769-78. doi: 10.1128/JB.01668-09. Epub 2010 Mar 26. J Bacteriol. 2010. PMID: 20348260 Free PMC article.
-
Positive regulation by nitrite of the nitrate assimilation operon in the cyanobacteria Synechococcus sp. strain PCC 7942 and Plectonema boryanum.J Bacteriol. 1996 Oct;178(19):5822-5. doi: 10.1128/jb.178.19.5822-5825.1996. J Bacteriol. 1996. PMID: 8824636 Free PMC article.
-
Role of NtcB in activation of nitrate assimilation genes in the cyanobacterium Synechocystis sp. strain PCC 6803.J Bacteriol. 2001 Oct;183(20):5840-7. doi: 10.1128/JB.183.20.5840-5847.2001. J Bacteriol. 2001. PMID: 11566981 Free PMC article.
-
Nitrate assimilation by bacteria.Adv Microb Physiol. 1998;39:1-30, 379. doi: 10.1016/s0065-2911(08)60014-4. Adv Microb Physiol. 1998. PMID: 9328645 Review.
-
Photosynthetic nitrate assimilation in cyanobacteria.Photosynth Res. 2005;83(2):117-33. doi: 10.1007/s11120-004-5830-9. Photosynth Res. 2005. PMID: 16143847 Review.
Cited by
-
Depicting Temporal, Functional, and Phylogenetic Patterns in Estuarine Diazotrophic Communities from Environmental DNA and RNA.Microb Ecol. 2021 Jan;81(1):36-51. doi: 10.1007/s00248-020-01562-1. Epub 2020 Aug 15. Microb Ecol. 2021. PMID: 32803362
-
Resistance to oxidative stress by inner membrane protein ElaB is regulated by OxyR and RpoS.Microb Biotechnol. 2019 Mar;12(2):392-404. doi: 10.1111/1751-7915.13369. Epub 2019 Jan 17. Microb Biotechnol. 2019. PMID: 30656833 Free PMC article.
-
Interaction of Type IV Toxin/Antitoxin Systems in Cryptic Prophages of Escherichia coli K-12.Toxins (Basel). 2017 Mar 1;9(3):77. doi: 10.3390/toxins9030077. Toxins (Basel). 2017. PMID: 28257056 Free PMC article.
-
sll1019 and slr1259 encoding glyoxalase II improve tolerance of Synechocystis sp. PCC 6803 to methylglyoxal- and ethanol- induced oxidative stress by glyoxalase pathway.Appl Environ Microbiol. 2024 Nov 20;90(11):e0056424. doi: 10.1128/aem.00564-24. Epub 2024 Oct 21. Appl Environ Microbiol. 2024. PMID: 39431850 Free PMC article.
-
Tail-Anchored Inner Membrane Protein ElaB Increases Resistance to Stress While Reducing Persistence in Escherichia coli.J Bacteriol. 2017 Apr 11;199(9):e00057-17. doi: 10.1128/JB.00057-17. Print 2017 May 1. J Bacteriol. 2017. PMID: 28242719 Free PMC article.
References
-
- Moreno-Vivián C, Flores E. 2007. Nitrate assimilation in bacteria, p 263–282. In Bothe H, Ferguson SJ, Newton WE (ed), Biology of the nitrogen cycle. Elsevier B.V., Amsterdam, Netherlands.

