Identifying personal microbiomes using metagenomic codes
- PMID: 25964341
- PMCID: PMC4460507
- DOI: 10.1073/pnas.1423854112
Identifying personal microbiomes using metagenomic codes
Abstract
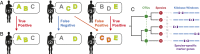
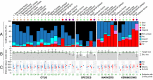
Community composition within the human microbiome varies across individuals, but it remains unknown if this variation is sufficient to uniquely identify individuals within large populations or stable enough to identify them over time. We investigated this by developing a hitting set-based coding algorithm and applying it to the Human Microbiome Project population. Our approach defined body site-specific metagenomic codes: sets of microbial taxa or genes prioritized to uniquely and stably identify individuals. Codes capturing strain variation in clade-specific marker genes were able to distinguish among 100s of individuals at an initial sampling time point. In comparisons with follow-up samples collected 30-300 d later, ∼30% of individuals could still be uniquely pinpointed using metagenomic codes from a typical body site; coincidental (false positive) matches were rare. Codes based on the gut microbiome were exceptionally stable and pinpointed >80% of individuals. The failure of a code to match its owner at a later time point was largely explained by the loss of specific microbial strains (at current limits of detection) and was only weakly associated with the length of the sampling interval. In addition to highlighting patterns of temporal variation in the ecology of the human microbiome, this work demonstrates the feasibility of microbiome-based identifiability-a result with important ethical implications for microbiome study design. The datasets and code used in this work are available for download from huttenhower.sph.harvard.edu/idability.
Keywords: forensic genetics; human microbiome; metagenomics; microbial ecology; strain variation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Comment in
-
Elucidating microbial codes to distinguish individuals.Proc Natl Acad Sci U S A. 2015 Jun 2;112(22):6778-9. doi: 10.1073/pnas.1507731112. Epub 2015 May 26. Proc Natl Acad Sci U S A. 2015. PMID: 26015565 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

