Optimized tools for multicolor stochastic labeling reveal diverse stereotyped cell arrangements in the fly visual system
- PMID: 25964354
- PMCID: PMC4460454
- DOI: 10.1073/pnas.1506763112
Optimized tools for multicolor stochastic labeling reveal diverse stereotyped cell arrangements in the fly visual system
Abstract
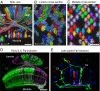
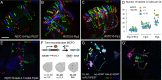
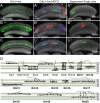
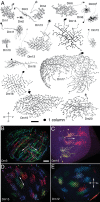
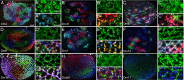
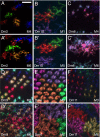
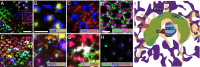
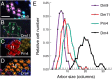
We describe the development and application of methods for high-throughput neuroanatomy in Drosophila using light microscopy. These tools enable efficient multicolor stochastic labeling of neurons at both low and high densities. Expression of multiple membrane-targeted and distinct epitope-tagged proteins is controlled both by a transcriptional driver and by stochastic, recombinase-mediated excision of transcription-terminating cassettes. This MultiColor FlpOut (MCFO) approach can be used to reveal cell shapes and relative cell positions and to track the progeny of precursor cells through development. Using two different recombinases, the number of cells labeled and the number of color combinations observed in those cells can be controlled separately. We demonstrate the utility of MCFO in a detailed study of diversity and variability of Distal medulla (Dm) neurons, multicolumnar local interneurons in the adult visual system. Similar to many brain regions, the medulla has a repetitive columnar structure that supports parallel information processing together with orthogonal layers of cell processes that enable communication between columns. We find that, within a medulla layer, processes of the cells of a given Dm neuron type form distinct patterns that reflect both the morphology of individual cells and the relative positions of their arbors. These stereotyped cell arrangements differ between cell types and can even differ for the processes of the same cell type in different medulla layers. This unexpected diversity of coverage patterns provides multiple independent ways of integrating visual information across the retinotopic columns and implies the existence of multiple developmental mechanisms that generate these distinct patterns.
Keywords: Drosophila; interneuron diversity; light microscopy; neuroanatomy; recombinase.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Masland RH. Neuronal diversity in the retina. Curr Opin Neurobiol. 2001;11(4):431–436. - PubMed
-
- Fischbach KF, Dittrich APM. The optic lobe of Drosophila melanogaster. 1. A Golgi analysis of wild-type structure. Cell Tissue Res. 1989;258(3):441–475.
-
- Grueber WB, Jan LY, Jan YN. Tiling of the Drosophila epidermis by multidendritic sensory neurons. Development. 2002;129(12):2867–2878. - PubMed
-
- Strausfeld NJ. Golgi studies on insects. 2. Optic lobes of Diptera. Philos T Roy Soc B. 1970;258(820):135. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

