Differences in long-term memory stability and AmCREB level between forward and backward conditioned honeybees (Apis mellifera)
- PMID: 25964749
- PMCID: PMC4410603
- DOI: 10.3389/fnbeh.2015.00091
Differences in long-term memory stability and AmCREB level between forward and backward conditioned honeybees (Apis mellifera)
Abstract
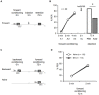
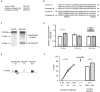
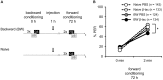
In classical conditioning a predictive relationship between a neutral stimulus (conditioned stimulus; CS) and a meaningful stimulus (unconditioned stimulus; US) is learned when the CS precedes the US. In backward conditioning the sequence of the stimuli is reversed. In this situation animals might learn that the CS signals the end or the absence of the US. In honeybees 30 min and 24 h following backward conditioning a memory for the excitatory and inhibitory properties of the CS could be retrieved, but it remains unclear whether a late long-term memory is formed that can be retrieved 72 h following backward conditioning. Here we examine this question by studying late long-term memory formation in forward and backward conditioning of the proboscis extension response (PER). We report a difference in the stability of memory formed upon forward and backward conditioning with the same number of conditioning trials. We demonstrate a transcription-dependent memory 72 h after forward conditioning but do not observe a 72 h memory after backward conditioning. Moreover we find that protein degradation is differentially involved in memory formation following these two conditioning protocols. We report differences in the level of a transcription factor, the cAMP response element binding protein (CREB) known to induce transcription underlying long-term memory formation, following forward and backward conditioning. Our results suggest that these alterations in CREB levels might be regulated by the proteasome. We propose that the differences observed are due to the sequence of stimulus presentation between forward and backward conditioning and not to differences in the strength of the association of both stimuli.
Keywords: CREB; backward conditioning; classical conditioning; long-term memory; proteasome; transcription; ubiquitin.
Figures


Similar articles
-
Short- and long-term memories formed upon backward conditioning in honeybees (Apis mellifera).Learn Mem. 2013 Dec 18;21(1):37-45. doi: 10.1101/lm.031765.113. Learn Mem. 2013. PMID: 24353291 Free PMC article.
-
Duration of the unconditioned stimulus in appetitive conditioning of honeybees differentially impacts learning, long-term memory strength, and the underlying protein synthesis.Learn Mem. 2014 Nov 17;21(12):676-85. doi: 10.1101/lm.035600.114. Print 2014 Dec. Learn Mem. 2014. PMID: 25403456 Free PMC article.
-
Forward and backward second-order Pavlovian conditioning in honeybees.Learn Mem. 2007 Oct 1;14(10):678-83. doi: 10.1101/lm.471307. Print 2007 Oct. Learn Mem. 2007. PMID: 17911371 Free PMC article.
-
Learning and memory in honeybees: from behavior to neural substrates.Annu Rev Neurosci. 1996;19:379-404. doi: 10.1146/annurev.ne.19.030196.002115. Annu Rev Neurosci. 1996. PMID: 8833448 Review.
-
Molecular mechanisms underlying formation of long-term reward memories and extinction memories in the honeybee (Apis mellifera).Learn Mem. 2014 Sep 15;21(10):534-42. doi: 10.1101/lm.033118.113. Print 2014 Oct. Learn Mem. 2014. PMID: 25225299 Free PMC article. Review.
Cited by
-
Involvement of phosphorylated Apis mellifera CREB in gating a honeybee's behavioral response to an external stimulus.Learn Mem. 2016 Apr 15;23(5):195-207. doi: 10.1101/lm.040964.115. Print 2016 May. Learn Mem. 2016. PMID: 27084927 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

