A distinct p53 target gene set predicts for response to the selective p53-HDM2 inhibitor NVP-CGM097
- PMID: 25965177
- PMCID: PMC4468608
- DOI: 10.7554/eLife.06498
A distinct p53 target gene set predicts for response to the selective p53-HDM2 inhibitor NVP-CGM097
Erratum in
-
Correction: A distinct p53 target gene set predicts for response to the selective p53-HDM2 inhibitor NVP-CGM097.Elife. 2016 Nov 17;5:e19317. doi: 10.7554/eLife.19317. Elife. 2016. PMID: 27852439 Free PMC article. No abstract available.
Abstract
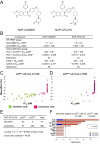
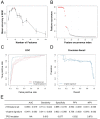
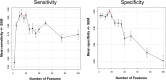
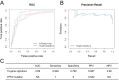
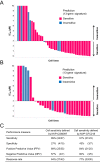
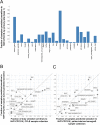
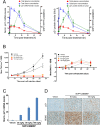
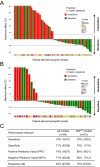
Biomarkers for patient selection are essential for the successful and rapid development of emerging targeted anti-cancer therapeutics. In this study, we report the discovery of a novel patient selection strategy for the p53-HDM2 inhibitor NVP-CGM097, currently under evaluation in clinical trials. By intersecting high-throughput cell line sensitivity data with genomic data, we have identified a gene expression signature consisting of 13 up-regulated genes that predicts for sensitivity to NVP-CGM097 in both cell lines and in patient-derived tumor xenograft models. Interestingly, these 13 genes are known p53 downstream target genes, suggesting that the identified gene signature reflects the presence of at least a partially activated p53 pathway in NVP-CGM097-sensitive tumors. Together, our findings provide evidence for the use of this newly identified predictive gene signature to refine the selection of patients with wild-type p53 tumors and increase the likelihood of response to treatment with p53-HDM2 inhibitors, such as NVP-CGM097.
Keywords: HDM2; human; human biology; medicine; p53; predictive signature; translational oncology.
Conflict of interest statement
SJ: Employee of Novartis Institutes for BioMedical Research.
SG: Employee of Novartis Institutes for BioMedical Research.
SF: Employee of Novartis Institutes for BioMedical Research.
HB: Employee of Novartis Institutes for BioMedical Research.
MI: Employee of Novartis Institutes for BioMedical Research.
TV: Employee of Novartis Institutes for BioMedical Research.
MM: Employee of Novartis Institutes for BioMedical Research.
SR: Employee of Novartis Institutes for BioMedical Research.
DAG: Employee of Novartis Institutes for BioMedical Research.
CR: Employee of Novartis Institutes for BioMedical Research.
MRJ: Employee of Novartis Institutes for BioMedical Research.
MW: Employee of Novartis Institutes for BioMedical Research.
JK: Employee of Novartis Institutes for BioMedical Research.
PF: Employee of Novartis Institutes for BioMedical Research.
FG: Employee of Novartis Institutes for BioMedical Research.
PH: Employee of Novartis Institutes for BioMedical Research.
KM: Employee of Novartis Institutes for BioMedical Research.
JW: Employee of Novartis Institutes for BioMedical Research.
EH: Employee of Novartis Institutes for BioMedical Research.
FH: Employee of Novartis Institutes for BioMedical Research.
WRS: Was an employee of Novartis Institutes for BioMedical Research and is now an employee of Peptidream Inc. and has ownership interest (including patents) in Peptidream Inc.
DGP: Employee of Novartis Institutes for BioMedical Research. Holds the position of VP/Global Head of Oncology in Novartis Institutes for BioMedical Research and has ownership interest (including patents) in Novartis Pharmaceuticals.
Figures


Comment in
-
A signature for success.Elife. 2015 Jun 16;4:e08773. doi: 10.7554/eLife.08773. Elife. 2015. PMID: 26079875 Free PMC article.
References
-
- Allen MA, Andrysik Z, Dengler VL, Mellert HS, Guarnieri A, Freeman JA, Sullivan KD, Galbraith MD, Luo X, Kraus WL, Dowell RD, Espinosa JM. Global analysis of p53-regulated transcription identifies its direct targets and unexpected regulatory mechanisms. eLife. 2014;3:e02200. doi: 10.7554/eLife.02200. - DOI - PMC - PubMed
-
- Anderson SJ, Lauritsen JP, Hartman MG, Foushee AM, Lefebvre JM, Shinton SA, Gerhardt B, Hardy RR, Oravecz T, Wiest DL. Ablation of ribosomal protein L22 selectively impairs alphabeta T cell development by activation of a p53-dependent checkpoint. Immunity. 2007;26:759–772. doi: 10.1016/j.immuni.2007.04.012. - DOI - PubMed
-
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehár J, Kryukov GV, Sonkin D, Reddy A, Liu M, Murray L, Berger MF, Monahan JE, Morais P, Meltzer J, Korejwa A, Jané-Valbuena J, Mapa FA, Thibault J, Bric-Furlong E, Raman P, Shipway A, Engels IH, Cheng J, Yu GK, Yu J, Aspesi P, de Silva M, Jagtap K, Jones MD, Wang L, Hatton C, Palescandolo E, Gupta S, Mahan S, Sougnez C, Onofrio RC, Liefeld T, MacConaill L, Winckler W, Reich M, Li N, Mesirov JP, Gabriel SB, Getz G, Ardlie K, Chan V, Myer VE, Weber BL, Porter J, Warmuth M, Finan P, Harris JL, Meyerson M, Golub TR, Morrissey MP, Sellers WR, Schlegel R, Garraway LA, Aspesi P. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

