Optimization of translation profiles enhances protein expression and solubility
- PMID: 25965266
- PMCID: PMC4428881
- DOI: 10.1371/journal.pone.0127039
Optimization of translation profiles enhances protein expression and solubility
Abstract
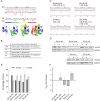
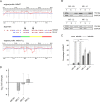
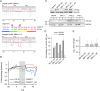
mRNA is translated with a non-uniform speed that actively coordinates co-translational folding of protein domains. Using structure-based homology we identified the structural domains in epoxide hydrolases (EHs) and introduced slow-translating codons to delineate the translation of single domains. These changes in translation speed dramatically improved the solubility of two EHs of metagenomic origin in Escherichia coli. Conversely, the importance of transient attenuation for the folding, and consequently solubility, of EH was evidenced with a member of the EH family from Agrobacterium radiobacter, which partitions in the soluble fraction when expressed in E. coli. Synonymous substitutions of codons shaping the slow-transiting regions to fast-translating codons render this protein insoluble. Furthermore, we show that low protein yield can be enhanced by decreasing the free folding energy of the initial 5'-coding region, which can disrupt mRNA secondary structure and enhance ribosomal loading. This study provides direct experimental evidence that mRNA is not a mere messenger for translation of codons into amino acids but bears an additional layer of information for folding, solubility and expression level of the encoded protein. Furthermore, it provides a general frame on how to modulate and fine-tune gene expression of a target protein.
Conflict of interest statement
Figures

Similar articles
-
Ribosome-mediated translational pause and protein domain organization.Protein Sci. 1996 Aug;5(8):1594-612. doi: 10.1002/pro.5560050814. Protein Sci. 1996. PMID: 8844849 Free PMC article.
-
Enantioconvergent production of (R)-1-phenyl-1,2-ethanediol from styrene oxide by combining the Solanum tuberosum and an evolved Agrobacterium radiobacter AD1 epoxide hydrolases.Biotechnol Bioeng. 2006 Jun 20;94(3):522-9. doi: 10.1002/bit.20860. Biotechnol Bioeng. 2006. PMID: 16498626
-
Identification of conserved slow codons that are important for protein expression and function.RNA Biol. 2021 Dec;18(12):2296-2307. doi: 10.1080/15476286.2021.1901185. Epub 2021 Apr 5. RNA Biol. 2021. PMID: 33691590 Free PMC article.
-
Timing during translation matters: synonymous mutations in human pathologies influence protein folding and function.Biochem Soc Trans. 2018 Aug 20;46(4):937-944. doi: 10.1042/BST20170422. Epub 2018 Jul 31. Biochem Soc Trans. 2018. PMID: 30065107 Review.
-
Synonymous Codons: Choose Wisely for Expression.Trends Genet. 2017 Apr;33(4):283-297. doi: 10.1016/j.tig.2017.02.001. Epub 2017 Mar 12. Trends Genet. 2017. PMID: 28292534 Free PMC article. Review.
Cited by
-
Synonymous and Nonsynonymous Substitutions in Dictyostelium discoideum Ammonium Transporter amtA Are Necessary for Functional Complementation in Saccharomyces cerevisiae.Microbiol Spectr. 2023 Feb 22;11(2):e0384722. doi: 10.1128/spectrum.03847-22. Online ahead of print. Microbiol Spectr. 2023. PMID: 36840598 Free PMC article.
-
Enhancing co-translational folding of heterologous protein by deleting non-essential ribosomal proteins in Pichia pastoris.Biotechnol Biofuels. 2019 Feb 21;12:38. doi: 10.1186/s13068-019-1377-z. eCollection 2019. Biotechnol Biofuels. 2019. PMID: 30828383 Free PMC article.
-
Pausing of Chloroplast Ribosomes Is Induced by Multiple Features and Is Linked to the Assembly of Photosynthetic Complexes.Plant Physiol. 2018 Mar;176(3):2557-2569. doi: 10.1104/pp.17.01564. Epub 2018 Jan 3. Plant Physiol. 2018. PMID: 29298822 Free PMC article.
-
Silent Polymorphisms: Can the tRNA Population Explain Changes in Protein Properties?Life (Basel). 2016 Feb 17;6(1):9. doi: 10.3390/life6010009. Life (Basel). 2016. PMID: 26901226 Free PMC article. Review.
-
Contribution of single amino acid and codon substitutions to the production and secretion of a lipase by Bacillus subtilis.Microb Cell Fact. 2017 Sep 25;16(1):160. doi: 10.1186/s12934-017-0772-z. Microb Cell Fact. 2017. PMID: 28946879 Free PMC article.
References
-
- Komili S, Silver PA. Coupling and coordination in gene expression processes: a systems biology view. Nat Rev Genet. 2008;9: 38–48. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

