Mutant U2AF1 Expression Alters Hematopoiesis and Pre-mRNA Splicing In Vivo
- PMID: 25965570
- PMCID: PMC4430854
- DOI: 10.1016/j.ccell.2015.04.008
Mutant U2AF1 Expression Alters Hematopoiesis and Pre-mRNA Splicing In Vivo
Abstract
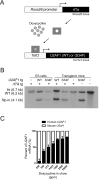
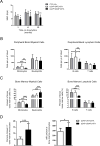
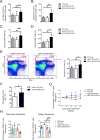
Heterozygous somatic mutations in the spliceosome gene U2AF1 occur in ∼ 11% of patients with myelodysplastic syndromes (MDS), the most common adult myeloid malignancy. It is unclear how these mutations contribute to disease. We examined in vivo hematopoietic consequences of the most common U2AF1 mutation using a doxycycline-inducible transgenic mouse model. Mice expressing mutant U2AF1(S34F) display altered hematopoiesis and changes in pre-mRNA splicing in hematopoietic progenitor cells by whole transcriptome analysis (RNA-seq). Integration with human RNA-seq datasets determined that common mutant U2AF1-induced splicing alterations are enriched in RNA processing genes, ribosomal genes, and recurrently mutated MDS and acute myeloid leukemia-associated genes. These findings support the hypothesis that mutant U2AF1 alters downstream gene isoform expression, thereby contributing to abnormal hematopoiesis in patients with MDS.
Copyright © 2015 Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

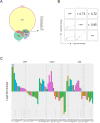
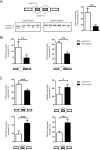
Comment in
-
Charting the "Splice" Routes to MDS.Cancer Cell. 2015 May 11;27(5):607-9. doi: 10.1016/j.ccell.2015.04.016. Cancer Cell. 2015. PMID: 25965565
References
-
- Bacher U, Haferlach T, Schnittger S, Zenger M, Meggendorfer M, Jeromin S, Roller A, Grossmann V, Krauth MT, Alpermann T, et al. Investigation of 305 patients with myelodysplastic syndromes and 20q deletion for associated cytogenetic and molecular genetic lesions and their prognostic impact. British journal of haematology. 2014;164:822–833. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

