Rhabdomyolysis-Associated Mutations in Human LPIN1 Lead to Loss of Phosphatidic Acid Phosphohydrolase Activity
- PMID: 25967228
- PMCID: PMC4484911
- DOI: 10.1007/8904_2015_440
Rhabdomyolysis-Associated Mutations in Human LPIN1 Lead to Loss of Phosphatidic Acid Phosphohydrolase Activity
Abstract
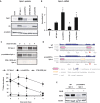
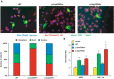
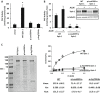
Rhabdomyolysis is an acute syndrome due to extensive injury of skeletal muscle. Recurrent rhabdomyolysis is often caused by inborn errors in intermediary metabolism, and recent work has suggested that mutations in the human gene encoding lipin 1 (LPIN1) may be a common cause of recurrent rhabdomyolysis in children. Lipin 1 dephosphorylates phosphatidic acid to form diacylglycerol (phosphatidic acid phosphohydrolase; PAP) and acts as a transcriptional regulatory protein to control metabolic gene expression. Herein, a 3-year-old boy with severe recurrent rhabdomyolysis was determined to be a compound heterozygote for a novel c.1904T>C (p.Leu635Pro) substitution and a previously reported genomic deletion of exons 18-19 (E766-S838_del) in LPIN1. Western blotting with patient muscle biopsy lysates demonstrated a marked reduction in lipin 1 protein, while immunohistochemical staining for lipin 1 showed abnormal subcellular localization. We cloned cDNAs to express recombinant lipin 1 proteins harboring pathogenic mutations and showed that the E766-S838_del allele was not expressed at the RNA or protein level. Lipin 1 p.Leu635Pro was expressed, but the protein was less stable, was aggregated in the cytosol, and was targeted for proteosomal degradation. Another pathogenic single amino acid substitution, lipin 1 p.Arg725His, was well expressed and retained its transcriptional regulatory function. However, both p.Leu635Pro and p.Arg725His proteins were found to be deficient in PAP activity. Kinetic analyses demonstrated a loss of catalysis rather than diminished substrate binding. These data suggest that loss of lipin 1-mediated PAP activity may be involved in the pathogenesis of rhabdomyolysis in lipin 1 deficiency.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

