Recovery from severe H7N9 disease is associated with diverse response mechanisms dominated by CD8⁺ T cells
- PMID: 25967273
- PMCID: PMC4479016
- DOI: 10.1038/ncomms7833
Recovery from severe H7N9 disease is associated with diverse response mechanisms dominated by CD8⁺ T cells
Abstract
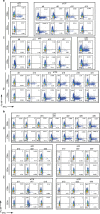
The avian origin A/H7N9 influenza virus causes high admission rates (>99%) and mortality (>30%), with ultimately favourable outcomes ranging from rapid recovery to prolonged hospitalization. Using a multicolour assay for monitoring adaptive and innate immunity, here we dissect the kinetic emergence of different effector mechanisms across the spectrum of H7N9 disease and recovery. We find that a diversity of response mechanisms contribute to resolution and survival. Patients discharged within 2-3 weeks have early prominent H7N9-specific CD8(+) T-cell responses, while individuals with prolonged hospital stays have late recruitment of CD8(+)/CD4(+) T cells and antibodies simultaneously (recovery by week 4), augmented even later by prominent NK cell responses (recovery >30 days). In contrast, those who succumbed have minimal influenza-specific immunity and little evidence of T-cell activation. Our study illustrates the importance of robust CD8(+) T-cell memory for protection against severe influenza disease caused by newly emerging influenza A viruses.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
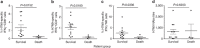




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

