Nuclear cyclophilins affect spliceosome assembly and function in vitro
- PMID: 25967372
- PMCID: PMC4537404
- DOI: 10.1042/BJ20150396
Nuclear cyclophilins affect spliceosome assembly and function in vitro
Abstract
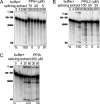
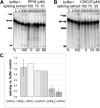
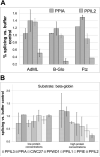
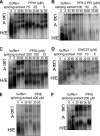
Cyclophilins are ubiquitously expressed proteins that bind to prolines and can catalyse cis/trans isomerization of proline residues. There are 17 annotated members of the cyclophilin family in humans, ubiquitously expressed and localized variously to the cytoplasm, nucleus or mitochondria. Surprisingly, all eight of the nuclear localized cyclophilins are found associated with spliceosomal complexes. However, their particular functions within this context are unknown. We have therefore adapted three established assays for in vitro pre-mRNA splicing to probe the functional roles of nuclear cyclophilins in the context of the human spliceosome. We find that four of the eight spliceosom-associated cyclophilins exert strong effects on splicing in vitro. These effects are dose-dependent and, remarkably, uniquely characteristic of each cyclophilin. Using both qualitative and quantitative means, we show that at least half of the nuclear cyclophilins can act as regulatory factors of spliceosome function in vitro. The present work provides the first quantifiable evidence that nuclear cyclophilins are splicing factors and provides a novel approach for future work into small molecule-based modulation of pre-mRNA splicing.
Keywords: nuclear cyclophilins; pre-messenger RNA (mRNA) splicing; spliceosome.
© 2015 Authors; published by Portland Press Limited.
Figures


References
-
- Wahl MC, Will CL, Lührmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. CrossRef PubMed. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

