Preclinical and clinical studies of the NEDD9 scaffold protein in cancer and other diseases
- PMID: 25967390
- PMCID: PMC4458429
- DOI: 10.1016/j.gene.2015.04.086
Preclinical and clinical studies of the NEDD9 scaffold protein in cancer and other diseases
Abstract
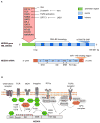
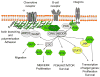
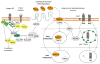
Cancer progression requires a significant reprogramming of cellular signaling to support the essential tumor-specific processes that include hyperproliferation, invasion (for solid tumors) and survival of metastatic colonies. NEDD9 (also known as CasL and HEF1) encodes a multi-domain scaffolding protein that assembles signaling complexes regulating multiple cellular processes relevant to cancer. These include responsiveness to signals emanating from the T and B cell receptors, integrins, chemokine receptors, and receptor tyrosine kinases, as well as cytoplasmic oncogenes such as BCR-ABL and FAK- and SRC-family kinases. Downstream, NEDD9 regulation of partners including CRKL, WAVE, PI3K/AKT, ERK, E-cadherin, Aurora-A (AURKA), HDAC6, and others allow NEDD9 to influence functions as pleiotropic as migration, invasion, survival, ciliary resorption, and mitosis. In this review, we summarize a growing body of preclinical and clinical data that indicate that while NEDD9 is itself non-oncogenic, changes in expression of NEDD9 (most commonly elevation of expression) are common features of tumors, and directly impact tumor aggressiveness, metastasis, and response to at least some targeted agents inhibiting NEDD9-interacting proteins. These data strongly support the relevance of further development of NEDD9 as a biomarker for therapeutic resistance. Finally, we briefly discuss emerging evidence supporting involvement of NEDD9 in additional pathological conditions, including stroke and polycystic kidney disease.
Keywords: CAS family protein; CAS-L; HEF1; Human malignancy; NEDD9; Prognosis; Protein function; Signal transduction.
Copyright © 2015 Elsevier B.V. All rights reserved.
Figures

References
-
- Kumar S, Tomooka Y, Noda M. Identification of a set of genes with developmentally down-regulated expression in the mouse brain. Biochem Biophys Res Commun. 1992;185(3):1155–61. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

