Reinforcement of STAT3 activity reprogrammes human embryonic stem cells to naive-like pluripotency
- PMID: 25968054
- PMCID: PMC4479042
- DOI: 10.1038/ncomms8095
Reinforcement of STAT3 activity reprogrammes human embryonic stem cells to naive-like pluripotency
Abstract
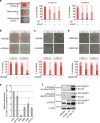
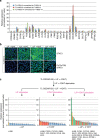
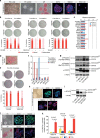
Leukemia inhibitory factor (LIF)/STAT3 signalling is a hallmark of naive pluripotency in rodent pluripotent stem cells (PSCs), whereas fibroblast growth factor (FGF)-2 and activin/nodal signalling is required to sustain self-renewal of human PSCs in a condition referred to as the primed state. It is unknown why LIF/STAT3 signalling alone fails to sustain pluripotency in human PSCs. Here we show that the forced expression of the hormone-dependent STAT3-ER (ER, ligand-binding domain of the human oestrogen receptor) in combination with 2i/LIF and tamoxifen allows human PSCs to escape from the primed state and enter a state characterized by the activation of STAT3 target genes and long-term self-renewal in FGF2- and feeder-free conditions. These cells acquire growth properties, a gene expression profile and an epigenetic landscape closer to those described in mouse naive PSCs. Together, these results show that temporarily increasing STAT3 activity is sufficient to reprogramme human PSCs to naive-like pluripotent cells.
Figures





References
-
- Evans M. J. & Kaufman M. H. Establishment in culture of pluripotential cells from mouse embryos. Nature 292, 154–156 (1981). - PubMed
-
- Smith A. G. et al. Inhibition of pluripotential embryonic stem cell differentiation by purified polypeptides. Nature 336, 688–690 (1988). - PubMed
-
- Tesar P. J. et al. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 448, 196–199 (2007). - PubMed
-
- Brons I. G. et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 448, 191–195 (2007). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

