Erratum to: DNA methylation age of human tissues and cell types
- PMID: 25968125
- PMCID: PMC4427927
- DOI: 10.1186/s13059-015-0649-6
Erratum to: DNA methylation age of human tissues and cell types
Figures
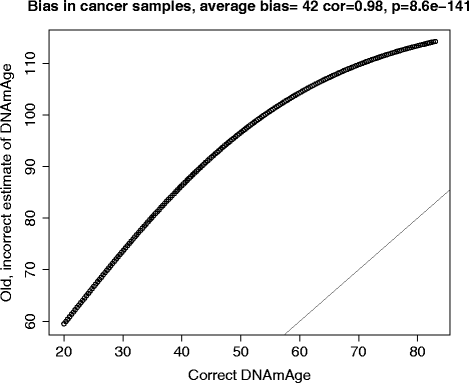
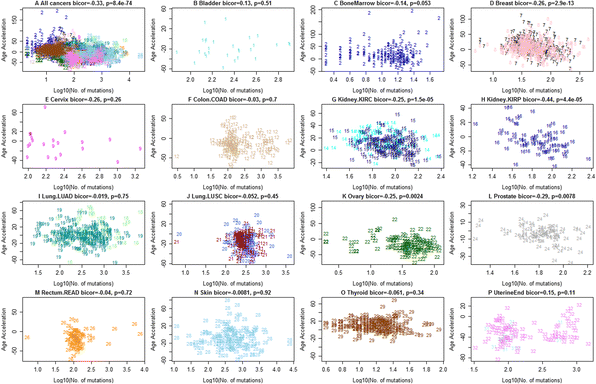
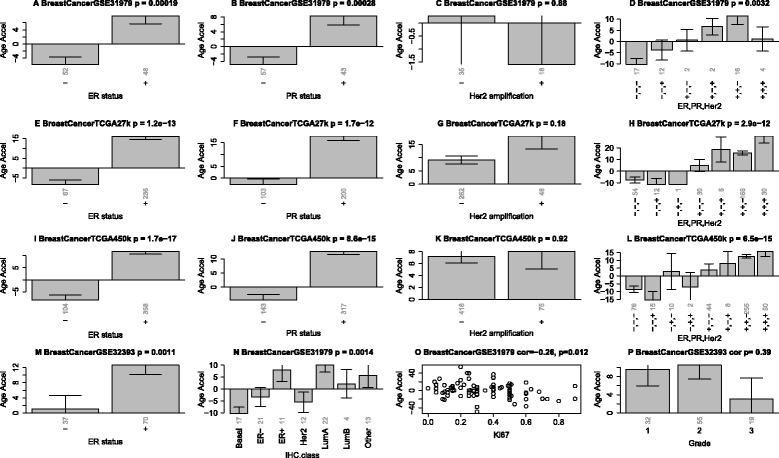
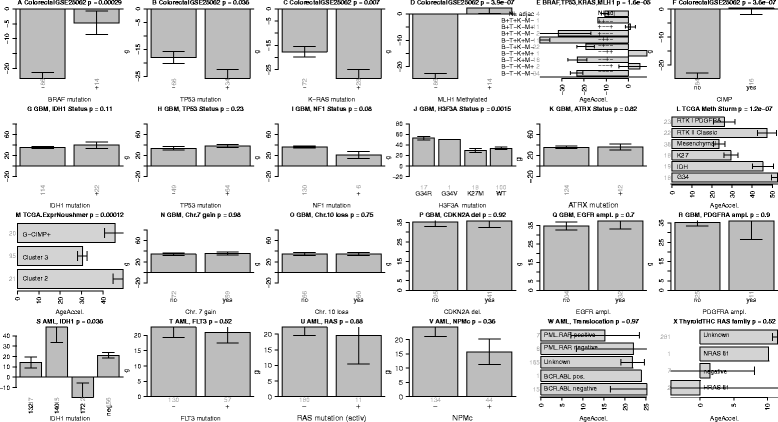
Erratum for
-
DNA methylation age of human tissues and cell types.Genome Biol. 2013;14(10):R115. doi: 10.1186/gb-2013-14-10-r115. Genome Biol. 2013. PMID: 24138928 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

