Utility of characters evolving at diverse rates of evolution to resolve quartet trees with unequal branch lengths: analytical predictions of long-branch effects
- PMID: 25968460
- PMCID: PMC4429678
- DOI: 10.1186/s12862-015-0364-7
Utility of characters evolving at diverse rates of evolution to resolve quartet trees with unequal branch lengths: analytical predictions of long-branch effects
Abstract
Background: The detection and avoidance of "long-branch effects" in phylogenetic inference represents a longstanding challenge for molecular phylogenetic investigations. A consequence of parallelism and convergence, long-branch effects arise in phylogenetic inference when there is unequal molecular divergence among lineages, and they can positively mislead inference based on parsimony especially, but also inference based on maximum likelihood and Bayesian approaches. Long-branch effects have been exhaustively examined by simulation studies that have compared the performance of different inference methods in specific model trees and branch length spaces.
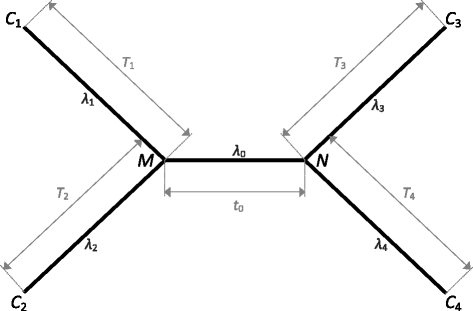
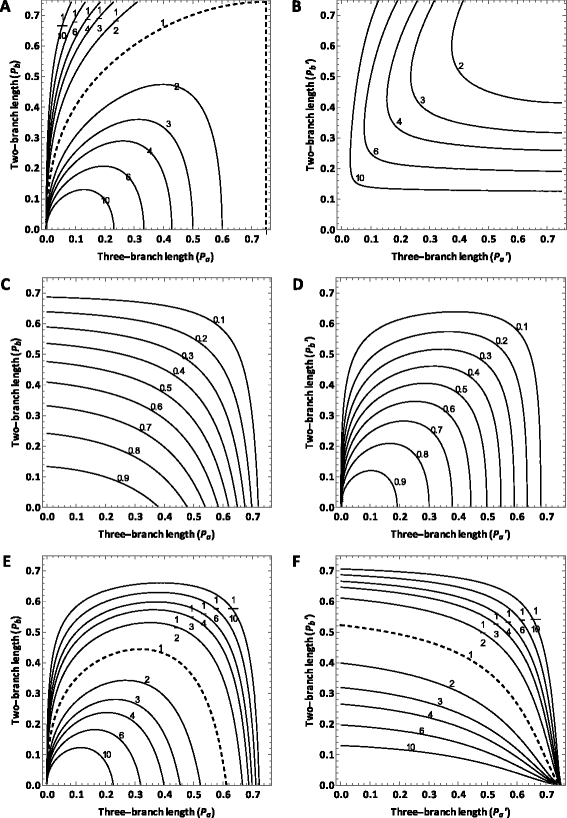
Results: In this paper, by generalizing the phylogenetic signal and noise analysis to quartets with uneven subtending branches, we quantify the utility of molecular characters for resolution of quartet phylogenies via parsimony. Our quantification incorporates contributions toward the correct tree from either signal or homoplasy (i.e. "the right result for either the right reason or the wrong reason"). We also characterize a highly conservative lower bound of utility that incorporates contributions to the correct tree only when they correspond to true, unobscured parsimony-informative sites (i.e. "the right result for the right reason"). We apply the generalized signal and noise analysis to classic quartet phylogenies in which long-branch effects can arise due to unequal rates of evolution or an asymmetrical topology. Application of the analysis leads to identification of branch length conditions in which inference will be inconsistent and reveals insights regarding how to improve sampling of molecular loci and taxa in order to correctly resolve phylogenies in which long-branch effects are hypothesized to exist.
Conclusions: The generalized signal and noise analysis provides analytical prediction of utility of characters evolving at diverse rates of evolution to resolve quartet phylogenies with unequal branch lengths. The analysis can be applied to identifying characters evolving at appropriate rates to resolve phylogenies in which long-branch effects are hypothesized to occur.
Figures




Similar articles
-
Bayesian and maximum likelihood phylogenetic analyses of protein sequence data under relative branch-length differences and model violation.BMC Evol Biol. 2005 Jan 28;5:8. doi: 10.1186/1471-2148-5-8. BMC Evol Biol. 2005. PMID: 15676079 Free PMC article.
-
Influence of long-branch bias on phylogenetic analysis.Genes Genet Syst. 2025 Mar 15;100. doi: 10.1266/ggs.24-00151. Epub 2025 Feb 15. Genes Genet Syst. 2025. PMID: 39956612
-
Reliable estimation of tree branch lengths using deep neural networks.PLoS Comput Biol. 2024 Aug 5;20(8):e1012337. doi: 10.1371/journal.pcbi.1012337. eCollection 2024 Aug. PLoS Comput Biol. 2024. PMID: 39102450 Free PMC article.
-
Stasis and convergence characterize morphological evolution in eupolypod II ferns.Ann Bot. 2014 Jan;113(1):35-54. doi: 10.1093/aob/mct247. Epub 2013 Nov 5. Ann Bot. 2014. PMID: 24197753 Free PMC article.
-
Statistical measures of uncertainty for branches in phylogenetic trees inferred from molecular sequences by using model-based methods.J Appl Genet. 2008;49(1):49-67. doi: 10.1007/BF03195249. J Appl Genet. 2008. PMID: 18263970 Review.
Cited by
-
Rubisco Adaptation Is More Limited by Phylogenetic Constraint Than by Catalytic Trade-off.Mol Biol Evol. 2021 Jun 25;38(7):2880-2896. doi: 10.1093/molbev/msab079. Mol Biol Evol. 2021. PMID: 33739416 Free PMC article.
-
Phylogenomic Subsampling and the Search for Phylogenetically Reliable Loci.Mol Biol Evol. 2021 Aug 23;38(9):4025-4038. doi: 10.1093/molbev/msab151. Mol Biol Evol. 2021. PMID: 33983409 Free PMC article.
-
Lineage-specific genes are clustered with HET-domain genes and respond to environmental and genetic manipulations regulating reproduction in Neurospora.PLoS Genet. 2023 Nov 7;19(11):e1011019. doi: 10.1371/journal.pgen.1011019. eCollection 2023 Nov. PLoS Genet. 2023. PMID: 37934795 Free PMC article.
-
PhyInformR: phylogenetic experimental design and phylogenomic data exploration in R.BMC Evol Biol. 2016 Dec 1;16(1):262. doi: 10.1186/s12862-016-0837-3. BMC Evol Biol. 2016. PMID: 27905871 Free PMC article.
-
Short branch attraction in phylogenomic inference under the multispecies coalescent.Front Ecol Evol. 2023;11:1134764. doi: 10.3389/fevo.2023.1134764. Epub 2023 Jun 28. Front Ecol Evol. 2023. PMID: 39233780 Free PMC article.
References
-
- Felsenstein J. Cases in which parsimony or compatibility methods will be positively misleading. Syst Zool. 1978;27:401–10. doi: 10.2307/2412923. - DOI
-
- Hendy MD, Penny D. A framework for the quantitative study of evolutionary trees. Syst Zool. 1989;38:297–309. doi: 10.2307/2992396. - DOI
-
- Kim JH. General inconsistency conditions for maximum parsimony: effects of branch lengths and increasing numbers of taxa. Syst Biol. 1996;45:363–74. doi: 10.1093/sysbio/45.3.363. - DOI
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

