Novel mechanism for scavenging of hypochlorite involving a periplasmic methionine-rich Peptide and methionine sulfoxide reductase
- PMID: 25968643
- PMCID: PMC4436054
- DOI: 10.1128/mBio.00233-15
Novel mechanism for scavenging of hypochlorite involving a periplasmic methionine-rich Peptide and methionine sulfoxide reductase
Abstract
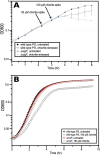
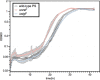
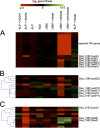
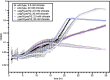
Reactive chlorine species (RCS) defense mechanisms are important for bacterial fitness in diverse environments. In addition to the anthropogenic use of RCS in the form of bleach, these compounds are also produced naturally through photochemical reactions of natural organic matter and in vivo by the mammalian immune system in response to invading microorganisms. To gain insight into bacterial RCS defense mechanisms, we investigated Azospira suillum strain PS, which produces periplasmic RCS as an intermediate of perchlorate respiration. Our studies identified an RCS response involving an RCS stress-sensing sigma/anti-sigma factor system (SigF/NrsF), a soluble hypochlorite-scavenging methionine-rich periplasmic protein (MrpX), and a putative periplasmic methionine sulfoxide reductase (YedY1). We investigated the underlying mechanism by phenotypic characterization of appropriate gene deletions, chemogenomic profiling of barcoded transposon pools, transcriptome sequencing, and biochemical assessment of methionine oxidation. Our results demonstrated that SigF was specifically activated by RCS and initiated the transcription of a small regulon centering around yedY1 and mrpX. A yedY1 paralog (yedY2) was found to have a similar fitness to yedY1 despite not being regulated by SigF. Markerless deletions of yedY2 confirmed its synergy with the SigF regulon. MrpX was strongly induced and rapidly oxidized by RCS, especially hypochlorite. Our results suggest a mechanism involving hypochlorite scavenging by sacrificial oxidation of the MrpX in the periplasm. Reduced MrpX is regenerated by the YedY methionine sulfoxide reductase activity. The phylogenomic distribution of this system revealed conservation in several Proteobacteria of clinical importance, including uropathogenic Escherichia coli and Brucella spp., implying a putative role in immune response evasion in vivo.
Importance: Bacteria are often stressed in the environment by reactive chlorine species (RCS) of either anthropogenic or natural origin, but little is known of the defense mechanisms they have evolved. Using a microorganism that generates RCS internally as part of its respiratory process allowed us to uncover a novel defense mechanism based on RCS scavenging by reductive reaction with a sacrificial methionine-rich peptide and redox recycling through a methionine sulfoxide reductase. This system is conserved in a broad diversity of organisms, including some of clinical importance, invoking a possible important role in innate immune system evasion.
Copyright © 2015 Melnyk et al.
Figures



References
-
- Keene WC, Khalil MAK, Erickson DJ, McCulloch A, Graedel TE, Lobert JM, Aucott ML, Gong SL, Harper DB, Kleiman G, Midgley P, Moore RM, Seuzaret C, Sturges WT, Benkovitz CM, Koropalov V, Barrie LA, Li YF. 1999. Composite global emissions of reactive chlorine from anthropogenic and natural sources: reactive chlorine emissions inventory. J Geophys Res 104:8429–8440. doi:10.1029/1998JD100084. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
