GalaxyPepDock: a protein-peptide docking tool based on interaction similarity and energy optimization
- PMID: 25969449
- PMCID: PMC4489314
- DOI: 10.1093/nar/gkv495
GalaxyPepDock: a protein-peptide docking tool based on interaction similarity and energy optimization
Abstract
Protein-peptide interactions are involved in a wide range of biological processes and are attractive targets for therapeutic purposes because of their small interfaces. Therefore, effective protein-peptide docking techniques can provide the basis for potential therapeutic applications by enabling an atomic-level understanding of protein interactions. With the increasing number of protein-peptide structures deposited in the protein data bank, the prediction accuracy of protein-peptide docking can be enhanced by utilizing the information provided by the database. The GalaxyPepDock web server, which is freely accessible at http://galaxy.seoklab.org/pepdock, performs similarity-based docking by finding templates from the database of experimentally determined structures and building models using energy-based optimization that allows for structural flexibility. The server can therefore effectively model the structural differences between the template and target protein-peptide complexes. The performance of GalaxyPepDock is superior to those of the other currently available web servers when tested on the PeptiDB set and on recently released complex structures. When tested on the CAPRI target 67, GalaxyPepDock generates models that are more accurate than the best server models submitted during the CAPRI blind prediction experiment.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

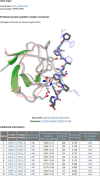
References
-
- Petsalaki E., Russell R.B. Peptide-mediated interactions in biological systems: new discoveries and applications. Curr. Opin. Biotechnol. 2008;19:344–350. - PubMed
-
- Wen W., Meinkoth J.L., Tsien R.Y., Taylor S.S. Identification of a signal for rapid export of proteins from the nucleus. Cell. 1995;82:463–473. - PubMed
-
- MacLaine N.J., Hupp T.R. How phosphorylation controls p53. Cell Cycle. 2011;10:916–921. - PubMed

