MSH3 Promotes Dynamic Behavior of Trinucleotide Repeat Tracts In Vivo
- PMID: 25969461
- PMCID: PMC4512540
- DOI: 10.1534/genetics.115.177303
MSH3 Promotes Dynamic Behavior of Trinucleotide Repeat Tracts In Vivo
Abstract
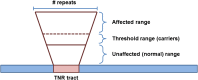
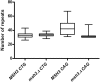
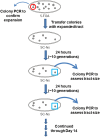
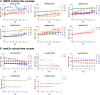
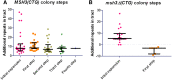
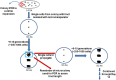
Trinucleotide repeat (TNR) expansions are the underlying cause of more than 40 neurodegenerative and neuromuscular diseases, including myotonic dystrophy and Huntington's disease, yet the pathway to expansion remains poorly understood. An important step in expansion is the shift from a stable TNR sequence to an unstable, expanding tract, which is thought to occur once a TNR attains a threshold length. Modeling of human data has indicated that TNR tracts are increasingly likely to expand as they increase in size and to do so in increments that are smaller than the repeat itself, but this has not been tested experimentally. Genetic work has implicated the mismatch repair factor MSH3 in promoting expansions. Using Saccharomyces cerevisiae as a model for CAG and CTG tract dynamics, we examined individual threshold-length TNR tracts in vivo over time in MSH3 and msh3Δ backgrounds. We demonstrate, for the first time, that these TNR tracts are highly dynamic. Furthermore, we establish that once such a tract has expanded by even a few repeat units, it is significantly more likely to expand again. Finally, we show that threshold- length TNR sequences readily accumulate net incremental expansions over time through a series of small expansion and contraction events. Importantly, the tracts were substantially stabilized in the msh3Δ background, with a bias toward contractions, indicating that Msh2-Msh3 plays an important role in shifting the expansion-contraction equilibrium toward expansion in the early stages of TNR tract expansion.
Keywords: Msh2-Msh3; Saccharomyces cerevisiae; mismatch repair; trinucleotide repeat tract.
Copyright © 2015 by the Genetics Society of America.
Figures




References
-
- Castel A. L., Cleary J. D., Pearson C. E., 2010. Repeat instability as the basis for human diseases and as a potential target for therapy. Nat. Rev. Mol. Cell Biol. 11: 165–170. - PubMed
-
- Concannon C., Lahue R. S., 2014. Nucleotide excision repair and the 26S proteasome function together to promote trinucleotide repeat expansions. DNA Repair 13: 42–49. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

