The effects of electronic medical record phenotyping details on genetic association studies: HDL-C as a case study
- PMID: 25969697
- PMCID: PMC4428098
- DOI: 10.1186/s13040-015-0048-2
The effects of electronic medical record phenotyping details on genetic association studies: HDL-C as a case study
Abstract
Background: Biorepositories linked to de-identified electronic medical records (EMRs) have the potential to complement traditional epidemiologic studies in genotype-phenotype studies of complex human diseases and traits. A major challenge in meeting this potential is the use of EMR-derived data to extract phenotypes and covariates for genetic association studies. Unlike traditional epidemiologic data, EMR-derived data are collected for clinical care and are therefore highly variable across patients. The variability of clinical data coupled with the challenges associated with searching unstructured clinical notes requires the development of algorithms to extract phenotypes for analysis. Given the number of possible algorithms that could be developed for any one EMR-derived phenotype, we explored here the impact algorithm decision logic has on genetic association study results for a single quantitative trait, high density lipoprotein cholesterol (HDL-C).
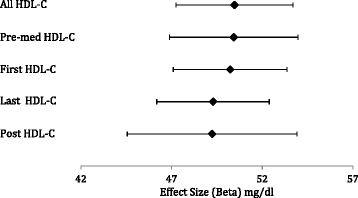
Results: We used five different algorithms to extract HDL-C from African American subjects genotyped on the Illumina Metabochip (n = 11,519) as part of Epidemiologic Architecture for Genes Linked to Environment (EAGLE). Tests of association between HDL-C and genetic risk scores for HDL-C associated variants suggest that the genetic effect size does not vary substantially across the five HDL-C definitions.
Conclusions: These data collectively suggest that, at least for this quantitative trait, algorithm decision logic and phenotyping details do not appreciably impact genetic association study test statistics.
Keywords: Electronic medical record; Genetic risk score; HDL-C; PAGE I study; eMERGE network.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

