Cooperative base pair melting by helicase and polymerase positioned one nucleotide from each other
- PMID: 25970034
- PMCID: PMC4460406
- DOI: 10.7554/eLife.06562
Cooperative base pair melting by helicase and polymerase positioned one nucleotide from each other
Abstract
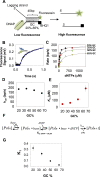
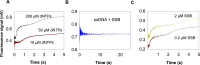
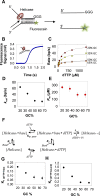
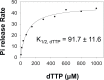
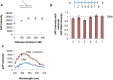
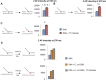
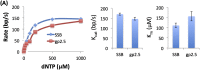
Leading strand DNA synthesis requires functional coupling between replicative helicase and DNA polymerase (DNAP) enzymes, but the structural and mechanistic basis of coupling is poorly understood. This study defines the precise positions of T7 helicase and T7 DNAP at the replication fork junction with single-base resolution to create a structural model that explains the mutual stimulation of activities. Our 2-aminopurine studies show that helicase and polymerase both participate in DNA melting, but each enzyme melts the junction base pair partially. When combined, the junction base pair is melted cooperatively provided the helicase is located one nucleotide ahead of the primer-end. The synergistic shift in equilibrium of junction base pair melting by combined enzymes explains the cooperativity, wherein helicase stimulates the polymerase by promoting dNTP binding (decreasing dNTP Km), polymerase stimulates the helicase by increasing the unwinding rate-constant (kcat), consequently the combined enzymes unwind DNA with kinetic parameters resembling enzymes translocating on single-stranded DNA.
Keywords: 2-aminopurine; DNA polymerase; E. coli; bacteriophage; biochemistry; biophysics; helicase; presteady state kinetics; replication; structural biology; viruses.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures








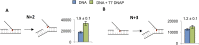


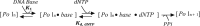



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

