Nonspecific yet decisive: Ubiquitination can affect the native-state dynamics of the modified protein
- PMID: 25970168
- PMCID: PMC4594657
- DOI: 10.1002/pro.2688
Nonspecific yet decisive: Ubiquitination can affect the native-state dynamics of the modified protein
Abstract
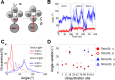
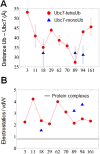
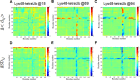
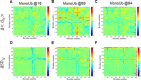
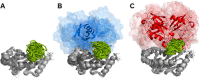
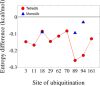
Ubiquitination is one of the most common post-translational modifications of proteins, and mediates regulated protein degradation among other cellular processes. A fundamental question regarding the mechanism of protein ubiquitination is whether and how ubiquitin affects the biophysical nature of the modified protein. For some systems, it was shown that the position of ubiquitin within the attachment site is quite flexible and ubiquitin does not specifically interact with its substrate. Nevertheless, it was revealed that polyubiquitination can decrease the thermal stability of the modified protein in a site-specific manner because of alterations of the thermodynamic properties of the folded and unfolded states. In this study, we used detailed atomistic simulations to focus on the molecular effects of ubiquitination on the native structure of the modified protein. As a model, we used Ubc7, which is an E2 enzyme whose in vivo ubiquitination process is well characterized and known to lead to degradation. We found that, despite the lack of specific direct interactions between the ubiquitin moiety and Ubc7, ubiquitination decreases the conformational flexibility of certain regions of the substrate Ubc7 protein, which reduces its entropy and thus destabilizes it. The strongest destabilizing effect was observed for systems in which Lys48-linked tetra-ubiquitin was attached to sites used for in vivo degradation. These results reveal how changes in the configurational entropy of the folded state may modulate the stability of the protein's native state. Overall, our results imply that ubiquitination can modify the biophysical properties of the attached protein in the folded state and that, in some proteins, different ubiquitination sites will lead to different biophysical outcomes. We propose that this destabilizing effect of polyubiquitin on the substrate is linked to the functions carried out by the modification, and in particular, regulatory control of protein half-life through proteasomal degradation.
Keywords: molecular dynamics simulations; multidomain protein; native state dynamics; ubiquitination.
© 2015 The Protein Society.
Figures




Similar articles
-
Mechanistic basis for ubiquitin modulation of a protein energy landscape.Proc Natl Acad Sci U S A. 2021 Mar 23;118(12):e2025126118. doi: 10.1073/pnas.2025126118. Proc Natl Acad Sci U S A. 2021. PMID: 33723075 Free PMC article.
-
Ubiquitin not only serves as a tag but also assists degradation by inducing protein unfolding.Proc Natl Acad Sci U S A. 2010 Feb 2;107(5):2001-6. doi: 10.1073/pnas.0912335107. Epub 2010 Jan 13. Proc Natl Acad Sci U S A. 2010. PMID: 20080694 Free PMC article.
-
The CUE Domain of Cue1 Aligns Growing Ubiquitin Chains with Ubc7 for Rapid Elongation.Mol Cell. 2016 Jun 16;62(6):918-928. doi: 10.1016/j.molcel.2016.04.031. Epub 2016 Jun 2. Mol Cell. 2016. PMID: 27264873
-
Use of E2~ubiquitin conjugates for the characterization of ubiquitin transfer by RING E3 ligases such as the inhibitor of apoptosis proteins.Methods Enzymol. 2014;545:243-63. doi: 10.1016/B978-0-12-801430-1.00010-X. Methods Enzymol. 2014. PMID: 25065893 Review.
-
Enzymes of ubiquitination and deubiquitination.Essays Biochem. 2012;52:37-50. doi: 10.1042/bse0520037. Essays Biochem. 2012. PMID: 22708562 Review.
Cited by
-
Site-specific ubiquitination affects protein energetics and proteasomal degradation.Nat Chem Biol. 2020 Aug;16(8):866-875. doi: 10.1038/s41589-020-0556-3. Epub 2020 Jun 1. Nat Chem Biol. 2020. PMID: 32483380 Free PMC article.
-
Design of genetically encoded sensors to detect nucleosome ubiquitination in live cells.J Cell Biol. 2021 Apr 5;220(4):e201911130. doi: 10.1083/jcb.201911130. J Cell Biol. 2021. PMID: 33570569 Free PMC article.
-
The Thermodynamic Properties of Fat10ylated Proteins Are Regulated by the Fat10ylation Site.ACS Omega. 2024 May 13;9(20):22265-22276. doi: 10.1021/acsomega.4c01396. eCollection 2024 May 21. ACS Omega. 2024. PMID: 38799324 Free PMC article.
-
Mechanistic basis for ubiquitin modulation of a protein energy landscape.Proc Natl Acad Sci U S A. 2021 Mar 23;118(12):e2025126118. doi: 10.1073/pnas.2025126118. Proc Natl Acad Sci U S A. 2021. PMID: 33723075 Free PMC article.
-
The Molten Globule State of a Globular Protein in a Cell Is More or Less Frequent Case Rather than an Exception.Molecules. 2022 Jul 7;27(14):4361. doi: 10.3390/molecules27144361. Molecules. 2022. PMID: 35889244 Free PMC article. Review.
References
-
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. - PubMed
-
- Varshavsky A. The ubiquitin system, an immense realm. Annu Rev Biochem. 2012;81:167–176. - PubMed
-
- Pickart CM, Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochim Biophys Acta. 2004;1695:55–72. - PubMed
-
- Chau V, Tobias JW, Bachmair A, Marriott D, Ecker DJ, Gonda DK, Varshavsky A. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science. 1989;243:1576–1583. - PubMed
-
- Chen ZJ, Sun LJ. Nonproteolytic functions of ubiquitin in cell signaling. Molecular Cell. 2009;33:275–286. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

