PPAR-α and glucocorticoid receptor synergize to promote erythroid progenitor self-renewal
- PMID: 25970251
- PMCID: PMC4498266
- DOI: 10.1038/nature14326
PPAR-α and glucocorticoid receptor synergize to promote erythroid progenitor self-renewal
Abstract
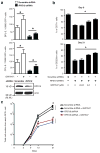
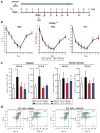
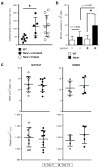
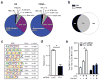
Many acute and chronic anaemias, including haemolysis, sepsis and genetic bone marrow failure diseases such as Diamond-Blackfan anaemia, are not treatable with erythropoietin (Epo), because the colony-forming unit erythroid progenitors (CFU-Es) that respond to Epo are either too few in number or are not sensitive enough to Epo to maintain sufficient red blood cell production. Treatment of these anaemias requires a drug that acts at an earlier stage of red cell formation and enhances the formation of Epo-sensitive CFU-E progenitors. Recently, we showed that glucocorticoids specifically stimulate self-renewal of an early erythroid progenitor, burst-forming unit erythroid (BFU-E), and increase the production of terminally differentiated erythroid cells. Here we show that activation of the peroxisome proliferator-activated receptor α (PPAR-α) by the PPAR-α agonists GW7647 and fenofibrate synergizes with the glucocorticoid receptor (GR) to promote BFU-E self-renewal. Over time these agonists greatly increase production of mature red blood cells in cultures of both mouse fetal liver BFU-Es and mobilized human adult CD34(+) peripheral blood progenitors, with a new and effective culture system being used for the human cells that generates normal enucleated reticulocytes. Although Ppara(-/-) mice show no haematological difference from wild-type mice in both normal and phenylhydrazine (PHZ)-induced stress erythropoiesis, PPAR-α agonists facilitate recovery of wild-type but not Ppara(-/-) mice from PHZ-induced acute haemolytic anaemia. We also show that PPAR-α alleviates anaemia in a mouse model of chronic anaemia. Finally, both in control and corticosteroid-treated BFU-E cells, PPAR-α co-occupies many chromatin sites with GR; when activated by PPAR-α agonists, additional PPAR-α is recruited to GR-adjacent sites and presumably facilitates GR-dependent BFU-E self-renewal. Our discovery of the role of PPAR-α agonists in stimulating self-renewal of early erythroid progenitor cells suggests that the clinically tested PPAR-α agonists we used may improve the efficacy of corticosteroids in treating Epo-resistant anaemias.
Conflict of interest statement
The authors declare no competing financial interests.
Figures









References
-
- Kaushansky KLAM, Beutler E, Kipps JT, Seligsohn U, Prchal TJ. Title of book: Williams Hematology. 8. 2010.
-
- Zimmerman JL. Use of blood products in sepsis: an evidence-based review. Critical care medicine. 2004;32:S542–547. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

