ChSeq: A database of chameleon sequences
- PMID: 25970262
- PMCID: PMC4500308
- DOI: 10.1002/pro.2689
ChSeq: A database of chameleon sequences
Abstract
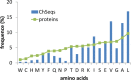
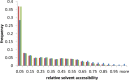
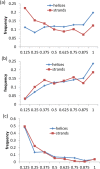
Chameleon sequences (ChSeqs) refer to sequence strings of identical amino acids that can adopt different conformations in protein structures. Researchers have detected and studied ChSeqs to understand the interplay between local and global interactions in protein structure formation. The different secondary structures adopted by one ChSeq challenge sequence-based secondary structure predictors. With increasing numbers of available Protein Data Bank structures, we here identify a large set of ChSeqs ranging from 6 to 10 residues in length. The homologous ChSeqs discovered highlight the structural plasticity involved in biological function. When compared with previous studies, the set of unrelated ChSeqs found represents an about 20-fold increase in the number of detected sequences, as well as an increase in the longest ChSeq length from 8 to 10 residues. We applied secondary structure predictors on our ChSeqs and found that methods based on a sequence profile outperformed methods based on a single sequence. For the unrelated ChSeqs, the evolutionary information provided by the sequence profile typically allows successful prediction of the prevailing secondary structure adopted in each protein family. Our dataset will facilitate future studies of ChSeqs, as well as interpretations of the interplay between local and nonlocal interactions. A user-friendly web interface for this ChSeq database is available at prodata.swmed.edu/chseq.
Keywords: ChSeq; biological function; chameleon sequence; conformational change; secondary structure; secondary structure prediction; sequence profile; structural plasticity.
© 2015 The Protein Society.
Figures
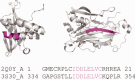
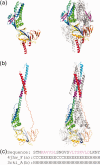



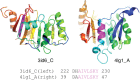
References
-
- Socci ND, Onuchic JN, Wolynes PG. Protein folding mechanisms and the multidimensional folding funnel. Proteins. 1998;32:136–158. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

