A Single Protein S-acyl Transferase Acts through Diverse Substrates to Determine Cryptococcal Morphology, Stress Tolerance, and Pathogenic Outcome
- PMID: 25970403
- PMCID: PMC4430228
- DOI: 10.1371/journal.ppat.1004908
A Single Protein S-acyl Transferase Acts through Diverse Substrates to Determine Cryptococcal Morphology, Stress Tolerance, and Pathogenic Outcome
Abstract
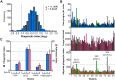
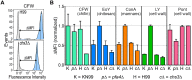
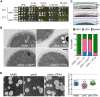
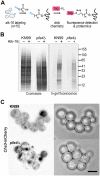
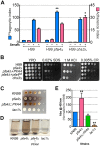
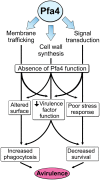
Cryptococcus neoformans is an opportunistic yeast that kills over 625,000 people yearly through lethal meningitis. Host phagocytes serve as the first line of defense against this pathogen, but fungal engulfment and subsequent intracellular proliferation also correlate with poor patient outcome. Defining the interactions of this facultative intracellular pathogen with host phagocytes is key to understanding the latter's opposing roles in infection and how they contribute to fungal latency, dissemination, and virulence. We used high-content imaging and a human monocytic cell line to screen 1,201 fungal mutants for strains with altered host interactions and identified multiple genes that influence fungal adherence and phagocytosis. One of these genes was PFA4, which encodes a protein S-acyl transferase (PAT), one of a family of DHHC domain-containing proteins that catalyzes lipid modification of proteins. Deletion of PFA4 caused dramatic defects in cryptococcal morphology, stress tolerance, and virulence. Bioorthogonal palmitoylome-profiling identified Pfa4-specific protein substrates involved in cell wall synthesis, signal transduction, and membrane trafficking responsible for these phenotypic alterations. We demonstrate that a single PAT is responsible for the modification of a subset of proteins that are critical in cryptococcal pathogenesis. Since several of these palmitoylated substrates are conserved in other pathogenic fungi, protein palmitoylation represents a potential avenue for new antifungal therapeutics.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
A sensitive high-throughput assay for evaluating host-pathogen interactions in Cryptococcus neoformans infection.PLoS One. 2011;6(7):e22773. doi: 10.1371/journal.pone.0022773. Epub 2011 Jul 28. PLoS One. 2011. PMID: 21829509 Free PMC article.
-
Sac1 links phosphoinositide turnover to cryptococcal virulence.mBio. 2024 Aug 14;15(8):e0149624. doi: 10.1128/mbio.01496-24. Epub 2024 Jul 2. mBio. 2024. PMID: 38953635 Free PMC article.
-
Aspartyl peptidase May1 induces host inflammatory response by altering cell wall composition in the fungal pathogen Cryptococcus neoformans.mBio. 2024 Jun 12;15(6):e0092024. doi: 10.1128/mbio.00920-24. Epub 2024 May 14. mBio. 2024. PMID: 38742885 Free PMC article.
-
Pathogenicity of Cryptococcus neoformans: virulence factors and immunological mechanisms.Microbes Infect. 1999 Apr;1(4):293-301. doi: 10.1016/s1286-4579(99)80025-2. Microbes Infect. 1999. PMID: 10602663 Review.
-
Sensing and responding to host-derived stress signals: lessons from fungal meningitis pathogen.Curr Opin Microbiol. 2024 Aug;80:102514. doi: 10.1016/j.mib.2024.102514. Epub 2024 Jul 18. Curr Opin Microbiol. 2024. PMID: 39024914 Review.
Cited by
-
The ER Protein Translocation Channel Subunit Sbh1 Controls Virulence of Cryptococcus neoformans.mBio. 2023 Feb 28;14(1):e0338422. doi: 10.1128/mbio.03384-22. Epub 2023 Feb 7. mBio. 2023. PMID: 36749043 Free PMC article.
-
Ergosterol distribution controls surface structure formation and fungal pathogenicity.mBio. 2023 Aug 31;14(4):e0135323. doi: 10.1128/mbio.01353-23. Epub 2023 Jul 6. mBio. 2023. PMID: 37409809 Free PMC article.
-
Protein Palmitoylation Plays an Important Role in Trichomonas vaginalis Adherence.Mol Cell Proteomics. 2018 Nov;17(11):2229-2241. doi: 10.1074/mcp.RA117.000018. Epub 2018 Feb 14. Mol Cell Proteomics. 2018. PMID: 29444981 Free PMC article.
-
A Family of Secretory Proteins Is Associated with Different Morphotypes in Cryptococcus neoformans.Appl Environ Microbiol. 2017 Feb 15;83(5):e02967-16. doi: 10.1128/AEM.02967-16. Print 2017 Mar 1. Appl Environ Microbiol. 2017. PMID: 28039134 Free PMC article.
-
Trojan Horse Transit Contributes to Blood-Brain Barrier Crossing of a Eukaryotic Pathogen.mBio. 2017 Jan 31;8(1):e02183-16. doi: 10.1128/mBio.02183-16. mBio. 2017. PMID: 28143979 Free PMC article.
References
-
- Bratton EW, El Husseini N, Chastain CA, Lee MS, Poole C, Sturmer T, et al. Comparison and temporal trends of three groups with cryptococcosis: HIV-infected, solid organ transplant, and HIV-negative/non-transplant. PLoS One. 2012;7(8):e43582 Epub 2012/09/01. 10.1371/journal.pone.0043582 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

