Conformational plasticity surrounding the active site of NADH oxidase from Thermus thermophilus
- PMID: 25970557
- PMCID: PMC4500311
- DOI: 10.1002/pro.2693
Conformational plasticity surrounding the active site of NADH oxidase from Thermus thermophilus
Abstract
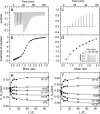
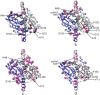
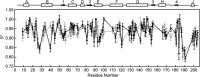
Biotechnological applications of enzymes can involve the use of these molecules under nonphysiological conditions. Thus, it is of interest to understand how environmental variables affect protein structure and dynamics and how this ultimately modulates enzyme function. NADH oxidase (NOX) from Thermus thermophilus exemplifies how enzyme activity can be tuned by reaction conditions, such as temperature, cofactor substitution, and the addition of cosolutes. This enzyme catalyzes the oxidation of reduced NAD(P)H to NAD(P)(+) with the concurrent reduction of O2 to H2O2, with relevance to biosensing applications. It is thermophilic, with an optimum temperature of approximately 65°C and sevenfold lower activity at 25°C. Moderate concentrations (≈1M) of urea and other chaotropes increase NOX activity by up to a factor of 2.5 at room temperature. Furthermore, it is a flavoprotein that accepts either FMN or the much larger FAD as cofactor. We have used nuclear magnetic resonance (NMR) titration and (15)N spin relaxation experiments together with isothermal titration calorimetry to study how NOX structure and dynamics are affected by changes in temperature, the addition of urea and the substitution of the FMN cofactor with FAD. The majority of signals from NOX are quite insensitive to changes in temperature, cosolute addition, and cofactor substitution. However, a small cluster of residues surrounding the active site shows significant changes. These residues are implicated in coupling changes in the solution conditions of the enzyme to changes in catalytic activity.
Keywords: NMR titration; enzyme cofactor substitution; isothermal titration calorimetry; nuclear magnetic resonance; spin relaxation; thermophile; urea activation.
© 2015 The Protein Society.
Figures



Similar articles
-
Crystal structure of NADH oxidase from Thermus thermophilus.Nat Struct Biol. 1995 Dec;2(12):1109-14. doi: 10.1038/nsb1295-1109. Nat Struct Biol. 1995. PMID: 8846223
-
A temperature-dependent conformational change of NADH oxidase from Thermus thermophilus HB8.Proteins. 2012 Feb;80(2):546-55. doi: 10.1002/prot.23219. Epub 2011 Nov 12. Proteins. 2012. PMID: 22081476 Free PMC article.
-
Active site dynamics in NADH oxidase from Thermus thermophilus studied by NMR spin relaxation.J Biomol NMR. 2011 Sep;51(1-2):71-82. doi: 10.1007/s10858-011-9542-0. Epub 2011 Sep 27. J Biomol NMR. 2011. PMID: 21947916
-
Enzyme-catalyzed and binding reaction kinetics determined by titration calorimetry.Biochim Biophys Acta. 2016 May;1860(5):957-966. doi: 10.1016/j.bbagen.2015.12.018. Epub 2015 Dec 22. Biochim Biophys Acta. 2016. PMID: 26721335 Review.
-
NO news is good news.Structure. 1998 Mar 15;6(3):255-8. doi: 10.1016/s0969-2126(98)00028-8. Structure. 1998. PMID: 9551547 Review.
Cited by
-
Perchlorate salts confer psychrophilic characteristics in α-chymotrypsin.Sci Rep. 2021 Aug 16;11(1):16523. doi: 10.1038/s41598-021-95997-2. Sci Rep. 2021. PMID: 34400699 Free PMC article.
-
The role of structural dynamics in the thermal adaptation of hyperthermophilic enzymes.Front Mol Biosci. 2022 Sep 7;9:981312. doi: 10.3389/fmolb.2022.981312. eCollection 2022. Front Mol Biosci. 2022. PMID: 36158582 Free PMC article.
-
Temperature dependence of NMR chemical shifts: Tracking and statistical analysis.Protein Sci. 2020 Jan;29(1):306-314. doi: 10.1002/pro.3785. Epub 2019 Nov 26. Protein Sci. 2020. PMID: 31730280 Free PMC article.
-
Functional Characterization and Structural Analysis of NADH Oxidase Mutants from Thermus thermophilus HB27: Role of Residues 166, 174, and 194 in the Catalytic Properties and Thermostability.Microorganisms. 2019 Oct 31;7(11):515. doi: 10.3390/microorganisms7110515. Microorganisms. 2019. PMID: 31683638 Free PMC article.
References
-
- Gianfreda L, Rao MA. Potential of extra cellular enzymes in remediation of polluted soils: a review. Enzyme Microb Technol. 2004;35:339–354.
-
- Panke S, Held M, Wubbolts M. Trends and innovations in industrial biocatalysis for the production of fine chemicals. Curr Opin Biotechnol. 2004;15:272–279. - PubMed
-
- Bistolas N, Wollenberger U, Jung C, Scheller FW. Cytochrome P450 biosensors—a review. Biosens Bioelectron. 2005;20:2408–2423. - PubMed
-
- Amine A, Mohammadi H, Bourais I, Palleschi G. Enzyme inhibition-based biosensors for food safety and environmental monitoring. Biosens Bioelectron. 2006;21:1405–1423. - PubMed
-
- Ahuja T, Mir IA, Kumar D, Rajesh Biomolecular immobilization on conducting polymers for biosensing applications. Biomaterials. 2007;28:791–805. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

