Patterns of gut bacterial colonization in three primate species
- PMID: 25970595
- PMCID: PMC4430486
- DOI: 10.1371/journal.pone.0124618
Patterns of gut bacterial colonization in three primate species
Abstract
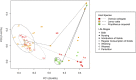
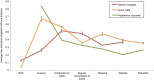
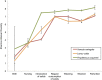
Host fitness is impacted by trillions of bacteria in the gastrointestinal tract that facilitate development and are inextricably tied to life history. During development, microbial colonization primes the gut metabolism and physiology, thereby setting the stage for adult nutrition and health. However, the ecological rules governing microbial succession are poorly understood. In this study, we examined the relationship between host lineage, captive diet, and life stage and gut microbiota characteristics in three primate species (infraorder, Lemuriformes). Fecal samples were collected from captive lemur mothers and their infants, from birth to weaning. Microbial DNA was extracted and the v4 region of 16S rDNA was sequenced on the Illumina platform using protocols from the Earth Microbiome Project. Here, we show that colonization proceeds along different successional trajectories in developing infants from species with differing dietary regimes and ecological profiles: frugivorous (fruit-eating) Varecia variegata, generalist Lemur catta, and folivorous (leaf-eating) Propithecus coquereli. Our analyses reveal community membership and succession patterns consistent with previous studies of human infants, suggesting that lemurs may serve as a useful model of microbial ecology in the primate gut. Each lemur species exhibits distinct species-specific bacterial diversity signatures correlating to life stages and life history traits, implying that gut microbial community assembly primes developing infants at species-specific rates for their respective adult feeding strategies.
Conflict of interest statement
Figures



References
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources

