Histone Sprocket Arginine Residues Are Important for Gene Expression, DNA Repair, and Cell Viability in Saccharomyces cerevisiae
- PMID: 25971662
- PMCID: PMC4512544
- DOI: 10.1534/genetics.115.175885
Histone Sprocket Arginine Residues Are Important for Gene Expression, DNA Repair, and Cell Viability in Saccharomyces cerevisiae
Abstract
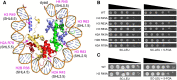
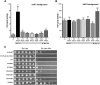
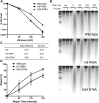
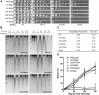
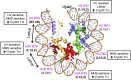
A critical feature of the intermolecular contacts that bind DNA to the histone octamer is the series of histone arginine residues that insert into the DNA minor groove at each superhelical location where the minor groove faces the histone octamer. One of these "sprocket" arginine residues, histone H4 R45, significantly affects chromatin structure in vivo and is lethal when mutated to alanine or cysteine in Saccharomyces cerevisiae (budding yeast). However, the roles of the remaining sprocket arginine residues (H3 R63, H3 R83, H2A R43, H2B R36, H2A R78, H3 R49) in chromatin structure and other cellular processes have not been well characterized. We have genetically characterized mutations in each of these histone residues when introduced either singly or in combination to yeast cells. We find that pairs of arginine residues that bind DNA adjacent to the DNA exit/entry sites in the nucleosome are lethal in yeast when mutated in combination and cause a defect in histone occupancy. Furthermore, mutations in individual residues compromise repair of UV-induced DNA lesions and affect gene expression and cryptic transcription. This study reveals simple rules for how the location and structural mode of DNA binding influence the biological function of each histone sprocket arginine residue.
Keywords: cryptic transcription; histone assembly; nucleosome; nucleotide excision repair.
Copyright © 2015 by the Genetics Society of America.
Figures

References
-
- Ahn S. H., Cheung W. L., Hsu J. Y., Diaz R. L., Smith M. M., et al. , 2005. Sterile 20 kinase phosphorylates histone H2B at serine 10 during hydrogen peroxide-induced apoptosis in S. cerevisiae. Cell 120: 25–36. - PubMed
-
- Bespalov V. A., Conconi A., Zhang X., Fahy D., Smerdon M. J., 2001. Improved method for measuring the ensemble average of strand breaks in genomic DNA. Environ. Mol. Mutagen. 38: 166–174. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

