Interaction of CK1δ with γTuSC ensures proper microtubule assembly and spindle positioning
- PMID: 25971801
- PMCID: PMC4571304
- DOI: 10.1091/mbc.E14-12-1627
Interaction of CK1δ with γTuSC ensures proper microtubule assembly and spindle positioning
Abstract
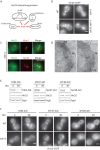
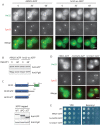
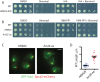
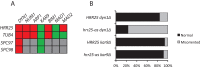
Casein kinase 1δ (CK1δ) family members associate with microtubule-organizing centers (MTOCs) from yeast to humans, but their mitotic roles and targets have yet to be identified. We show here that budding yeast CK1δ, Hrr25, is a γ-tubulin small complex (γTuSC) binding factor. Moreover, Hrr25's association with γTuSC depends on its kinase activity and its noncatalytic central domain. Loss of Hrr25 kinase activity resulted in assembly of unusually long cytoplasmic microtubules and defects in spindle positioning, consistent with roles in regulation of γTuSC-mediated microtubule nucleation and the Kar9 spindle-positioning pathway, respectively. Hrr25 directly phosphorylated γTuSC proteins in vivo and in vitro, and this phosphorylation promoted γTuSC integrity and activity. Because CK1δ and γTuSC are highly conserved and present at MTOCs in diverse eukaryotes, similar regulatory mechanisms are expected to apply generally in eukaryotes.
© 2015 Peng et al. This article is distributed by The American Society for Cell Biology under license from the author(s). Two months after publication it is available to the public under an Attribution–Noncommercial–Share Alike 3.0 Unported Creative Commons License (http://creativecommons.org/licenses/by-nc-sa/3.0).
Figures

Similar articles
-
Higher-order oligomerization of Spc110p drives γ-tubulin ring complex assembly.Mol Biol Cell. 2016 Jul 15;27(14):2245-58. doi: 10.1091/mbc.E16-02-0072. Epub 2016 May 25. Mol Biol Cell. 2016. PMID: 27226487 Free PMC article.
-
Microtubule nucleation at non-spindle pole body microtubule-organizing centers requires fission yeast centrosomin-related protein mod20p.Curr Biol. 2004 May 4;14(9):763-75. doi: 10.1016/j.cub.2004.03.042. Curr Biol. 2004. PMID: 15120067
-
Gamma-tubulin is required for proper recruitment and assembly of Kar9-Bim1 complexes in budding yeast.Mol Biol Cell. 2006 Oct;17(10):4420-34. doi: 10.1091/mbc.e06-03-0245. Epub 2006 Aug 9. Mol Biol Cell. 2006. PMID: 16899509 Free PMC article.
-
Microtubule nucleation: gamma-tubulin and beyond.J Cell Sci. 2006 Oct 15;119(Pt 20):4143-53. doi: 10.1242/jcs.03226. J Cell Sci. 2006. PMID: 17038541 Review.
-
Targeting of γ-tubulin complexes to microtubule organizing centers: conservation and divergence.Trends Cell Biol. 2015 May;25(5):296-307. doi: 10.1016/j.tcb.2014.12.002. Epub 2014 Dec 24. Trends Cell Biol. 2015. PMID: 25544667 Review.
Cited by
-
CK1-mediated phosphorylation of FAM110A promotes its interaction with mitotic spindle and controls chromosomal alignment.EMBO Rep. 2021 Jul 5;22(7):e51847. doi: 10.15252/embr.202051847. Epub 2021 May 26. EMBO Rep. 2021. PMID: 34080749 Free PMC article.
-
Mitotic spindle formation in the absence of Polo kinase.Proc Natl Acad Sci U S A. 2022 Mar 22;119(12):e2114429119. doi: 10.1073/pnas.2114429119. Epub 2022 Mar 14. Proc Natl Acad Sci U S A. 2022. PMID: 35286199 Free PMC article.
-
Casein Kinase I Protein Hrr25 Is Required for Pin4 Phosphorylation and Mediates Cell Wall Integrity Signaling in Saccharomyces cerevisiae.Genes (Basel). 2025 Jan 17;16(1):94. doi: 10.3390/genes16010094. Genes (Basel). 2025. PMID: 39858641 Free PMC article.
-
Structure of the Saccharomyces cerevisiae Hrr25:Mam1 monopolin subcomplex reveals a novel kinase regulator.EMBO J. 2016 Oct 4;35(19):2139-2151. doi: 10.15252/embj.201694082. Epub 2016 Aug 4. EMBO J. 2016. PMID: 27491543 Free PMC article.
-
CK1 Is a Druggable Regulator of Microtubule Dynamics and Microtubule-Associated Processes.Cancers (Basel). 2022 Mar 5;14(5):1345. doi: 10.3390/cancers14051345. Cancers (Basel). 2022. PMID: 35267653 Free PMC article. Review.
References
-
- Aldaz H, Rice LM, Stearns T, Agard DA. Insights into microtubule nucleation from the crystal structure of human gamma-tubulin. Nature. 2005;435:523–527. - PubMed
-
- Alvarado-Kristensson M, Rodriguez MJ, Silio V, Valpuesta JM, Carrera AC. SADB phosphorylation of gamma-tubulin regulates centrosome duplication. Nat Cell Biol. 2009;11:1081–1092. - PubMed
-
- Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426:570–574. - PubMed
-
- Bahtz R, Seidler J, Arnold M, Haselmann-Weiss U, Antony C, Lehmann WD, Hoffmann I. GCP6 is a substrate of Plk4 and required for centriole duplication. J Cell Sci. 2012;125:486–496. - PubMed
-
- Behrend L, Stoter M, Kurth M, Rutter G, Heukeshoven J, Deppert W, Knippschild U. Interaction of casein kinase 1 delta (CK1delta) with post-Golgi structures, microtubules and the spindle apparatus. Eur J Cell Biol. 2000;79:240–251. - PubMed

