A novel tropomyosin isoform functions at the mitotic spindle and Golgi in Drosophila
- PMID: 25971803
- PMCID: PMC4571303
- DOI: 10.1091/mbc.E14-12-1619
A novel tropomyosin isoform functions at the mitotic spindle and Golgi in Drosophila
Abstract
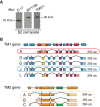
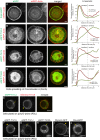
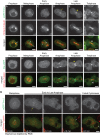
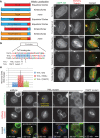
Most eukaryotic cells express multiple isoforms of the actin-binding protein tropomyosin that help construct a variety of cytoskeletal networks. Only one nonmuscle tropomyosin (Tm1A) has previously been described in Drosophila, but developmental defects caused by insertion of P-elements near tropomyosin genes imply the existence of additional, nonmuscle isoforms. Using biochemical and molecular genetic approaches, we identified three tropomyosins expressed in Drosophila S2 cells: Tm1A, Tm1J, and Tm2A. The Tm1A isoform localizes to the cell cortex, lamellar actin networks, and the cleavage furrow of dividing cells--always together with myosin-II. Isoforms Tm1J and Tm2A colocalize around the Golgi apparatus with the formin-family protein Diaphanous, and loss of either isoform perturbs cell cycle progression. During mitosis, Tm1J localizes to the mitotic spindle, where it promotes chromosome segregation. Using chimeras, we identified the determinants of tropomyosin localization near the C-terminus. This work 1) identifies and characterizes previously unknown nonmuscle tropomyosins in Drosophila, 2) reveals a function for tropomyosin in the mitotic spindle, and 3) uncovers sequence elements that specify isoform-specific localizations and functions of tropomyosin.
© 2015 Goins and Mullins. This article is distributed by The American Society for Cell Biology under license from the author(s). Two months after publication it is available to the public under an Attribution–Noncommercial–Share Alike 3.0 Unported Creative Commons License (http://creativecommons.org/licenses/by-nc-sa/3.0).
Figures



Similar articles
-
Myosin motor isoforms direct specification of actomyosin function by tropomyosins.Cytoskeleton (Hoboken). 2015 Mar;72(3):131-45. doi: 10.1002/cm.21213. Epub 2015 Mar 26. Cytoskeleton (Hoboken). 2015. PMID: 25712463 Free PMC article.
-
Differential regulation of skeletal muscle myosin-II and brush border myosin-I enzymology and mechanochemistry by bacterially produced tropomyosin isoforms.Cell Motil Cytoskeleton. 1994;29(1):29-45. doi: 10.1002/cm.970290104. Cell Motil Cytoskeleton. 1994. PMID: 7820856
-
Moesin controls cortical rigidity, cell rounding, and spindle morphogenesis during mitosis.Curr Biol. 2008 Jan 22;18(2):91-101. doi: 10.1016/j.cub.2007.12.051. Curr Biol. 2008. PMID: 18207738
-
[Role of tropomyosin isoforms in diversification of actin filaments functions].Postepy Biochem. 2009;55(2):201-6. Postepy Biochem. 2009. PMID: 19824477 Review. Polish.
-
Tropomyosin isoforms in nonmuscle cells.Int Rev Cytol. 1997;170:1-38. doi: 10.1016/s0074-7696(08)61619-8. Int Rev Cytol. 1997. PMID: 9002235 Review.
Cited by
-
On-target action of anti-tropomyosin drugs regulates glucose metabolism.Sci Rep. 2018 Mar 15;8(1):4604. doi: 10.1038/s41598-018-22946-x. Sci Rep. 2018. PMID: 29545590 Free PMC article.
-
Parallel assembly of actin and tropomyosin, but not myosin II, during de novo actin filament formation in live mice.J Cell Sci. 2018 Mar 19;131(6):jcs212654. doi: 10.1242/jcs.212654. J Cell Sci. 2018. PMID: 29487177 Free PMC article.
-
Targeting the cytoskeleton against metastatic dissemination.Cancer Metastasis Rev. 2021 Mar;40(1):89-140. doi: 10.1007/s10555-020-09936-0. Epub 2021 Jan 20. Cancer Metastasis Rev. 2021. PMID: 33471283 Review.
-
Actin-tropomyosin distribution in non-muscle cells.J Muscle Res Cell Motil. 2020 Mar;41(1):11-22. doi: 10.1007/s10974-019-09514-0. Epub 2019 May 4. J Muscle Res Cell Motil. 2020. PMID: 31054005 Free PMC article. Review.
-
An Atypical Tropomyosin in Drosophila with Intermediate Filament-like Properties.Cell Rep. 2016 Jul 26;16(4):928-938. doi: 10.1016/j.celrep.2016.06.054. Epub 2016 Jul 7. Cell Rep. 2016. PMID: 27396338 Free PMC article.
References
-
- Bettencourt-Dias M, Goshima G. RNAi in Drosophila S2 cells as a tool for studying cell cycle progression. Methods Mol Biol. 2009;545:39–62. - PubMed
-
- Blanchoin L, Pollard TD, Hitchcock-DeGregori SE. Inhibition of the Arp2/3 complex-nucleated actin polymerization and branch formation by tropomyosin. Curr Biol. 2001;11:1300–1304. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

