Light-induced Changes in the Dimerization Interface of Bacteriophytochromes
- PMID: 25971964
- PMCID: PMC4481235
- DOI: 10.1074/jbc.M115.650127
Light-induced Changes in the Dimerization Interface of Bacteriophytochromes
Abstract
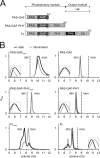
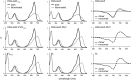
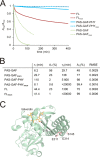
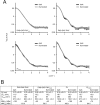
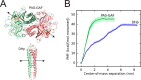
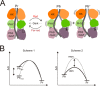
Phytochromes are dimeric photoreceptor proteins that sense red light levels in plants, fungi, and bacteria. The proteins are structurally divided into a light-sensing photosensory module consisting of PAS, GAF, and PHY domains and a signaling output module, which in bacteriophytochromes typically is a histidine kinase (HK) domain. Existing structural data suggest that two dimerization interfaces exist between the GAF and HK domains, but their functional roles remain unclear. Using mutational, biochemical, and computational analyses of the Deinococcus radiodurans phytochrome, we demonstrate that two dimerization interfaces between sister GAF and HK domains stabilize the dimer with approximately equal contributions. The existence of both dimerization interfaces is critical for thermal reversion back to the resting state. We also find that a mutant in which the interactions between the GAF domains were removed monomerizes under red light. This implies that the interactions between the HK domains are significantly altered by photoconversion. The results suggest functional importance of the dimerization interfaces in bacteriophytochromes.
Keywords: cell signaling; dimerization; high performance liquid chromatography (HPLC); histidine kinase; molecular dynamics; mutagenesis; photoreceptor; phytochrome; protein conformation; x-ray scattering.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures
References
-
- Quail P. H. (2002) Phytochrome photosensory signalling networks. Nat. Rev. Mol. Cell Biol. 3, 85–93 - PubMed
-
- Krieger A., Molina I., Oberpichler I., Michael N., Lamparter T. (2008) Spectral properties of phytochrome Agp2 from Agrobacterium tumefaciens are specifically modified by a compound of the cell extract. J. Photochem. Photobiol. B. 93, 16–22 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

