Computation and Functional Studies Provide a Model for the Structure of the Zinc Transporter hZIP4
- PMID: 25971965
- PMCID: PMC4505028
- DOI: 10.1074/jbc.M114.617613
Computation and Functional Studies Provide a Model for the Structure of the Zinc Transporter hZIP4
Abstract
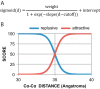
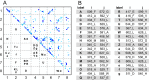
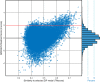
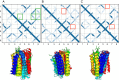
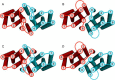
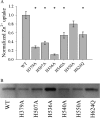
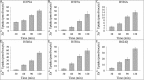
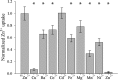
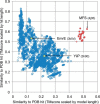
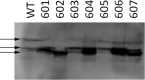
Members of the Zrt and Irt protein (ZIP) family are a central participant in transition metal homeostasis as they function to increase the cytosolic concentration of zinc and/or iron. However, the lack of a crystal structure hinders elucidation of the molecular mechanism of ZIP proteins. Here, we employed GREMLIN, a co-evolution-based contact prediction approach in conjunction with the Rosetta structure prediction program to construct a structural model of the human (h) ZIP4 transporter. The predicted contact data are best fit by modeling hZIP4 as a dimer. Mutagenesis of residues that comprise a central putative hZIP4 transmembrane transition metal coordination site in the structural model alter the kinetics and specificity of hZIP4. Comparison of the hZIP4 dimer model to all known membrane protein structures identifies the 12-transmembrane monomeric Piriformospora indica phosphate transporter (PiPT), a member of the major facilitator superfamily (MFS), as a likely structural homolog.
Keywords: computer modeling; membrane biophysics; membrane protein; metal homeostasis; metal ion-protein interaction; protein evolution; transport metal; transporter; zinc.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures




References
-
- Eide D. J. (2004) The SLC39 family of metal ion transporters. Pflugers Arch. 447, 796–800 - PubMed
-
- Dempski R. E. (2012) The cation selectivity of the ZIP transporters. Curr. Top. Membr. 69, 221–245 - PubMed
-
- Antala S., Dempski R. E. (2012) The human ZIP4 transporter has two distinct binding affinities and mediates transport of multiple transition metals. Biochemistry 51, 963–973 - PubMed
-
- Pinilla-Tenas J. J., Sparkman B. K., Shawki A., Illing A. C., Mitchell C. J., Zhao N., Liuzzi J. P., Cousins R. J., Knutson M. D., Mackenzie B. (2011) Zip14 is a complex broad-scope metal-ion transporter whose functional properties support roles in the cellular uptake of zinc and nontransferrin-bound iron. Am. J. Physiol. Cell Physiol. 301, C862–C871 - PMC - PubMed
-
- Gaither L. A., Eide D. J. (2001) Eukaryotic zinc transporters and their regulation. Biometals 14, 251–270 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

