Functional inflammatory profiles distinguish myelin-reactive T cells from patients with multiple sclerosis
- PMID: 25972006
- PMCID: PMC4497538
- DOI: 10.1126/scitranslmed.aaa8038
Functional inflammatory profiles distinguish myelin-reactive T cells from patients with multiple sclerosis
Abstract
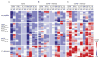
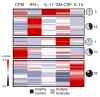
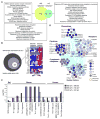
Myelin-reactive T cells have been identified in patients with multiple sclerosis (MS) and healthy subjects with comparable frequencies, but the contribution of these autoreactive T cells to disease pathology remains unknown. A total of 13,324 T cell libraries generated from blood of 23 patients and 22 healthy controls were interrogated for reactivity to myelin antigens. Libraries derived from CCR6(+) myelin-reactive T cells from patients with MS exhibited significantly enhanced production of interferon-γ (IFN-γ), interleukin-17 (IL-17), and granulocyte-macrophage colony-stimulating factor (GM-CSF) compared to healthy controls. Single-cell clones isolated by major histocompatibility complex/peptide tetramers from CCR6(+) T cell libraries also secreted more proinflammatory cytokines, whereas clones isolated from controls secreted more IL-10. The transcriptomes of myelin-specific CCR6(+) T cells from patients with MS were distinct from those derived from healthy controls and, notably, were enriched in T helper cell 17 (TH17)-induced experimental autoimmune encephalitis gene signatures, and gene signatures derived from TH17 cells isolated other human autoimmune diseases. These data, although not causal, imply that functional differences between antigen-specific T cells from MS and healthy controls are fundamental to disease development and support the notion that IL-10 production from myelin-reactive T cells may act to limit disease progression or even pathogenesis.
Copyright © 2015, American Association for the Advancement of Science.
Conflict of interest statement
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

