Rapid Whole-Genome Sequencing of Mycobacterium tuberculosis Isolates Directly from Clinical Samples
- PMID: 25972414
- PMCID: PMC4473240
- DOI: 10.1128/JCM.00486-15
Rapid Whole-Genome Sequencing of Mycobacterium tuberculosis Isolates Directly from Clinical Samples
Abstract
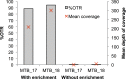
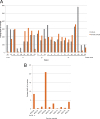
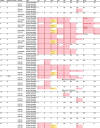
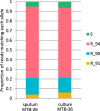
The rapid identification of antimicrobial resistance is essential for effective treatment of highly resistant Mycobacterium tuberculosis. Whole-genome sequencing provides comprehensive data on resistance mutations and strain typing for monitoring transmission, but unlike for conventional molecular tests, this has previously been achievable only from cultures of M. tuberculosis. Here we describe a method utilizing biotinylated RNA baits designed specifically for M. tuberculosis DNA to capture full M. tuberculosis genomes directly from infected sputum samples, allowing whole-genome sequencing without the requirement of culture. This was carried out on 24 smear-positive sputum samples, collected from the United Kingdom and Lithuania where a matched culture sample was available, and 2 samples that had failed to grow in culture. M. tuberculosis sequencing data were obtained directly from all 24 smear-positive culture-positive sputa, of which 20 were of high quality (>20× depth and >90% of the genome covered). Results were compared with those of conventional molecular and culture-based methods, and high levels of concordance between phenotypical resistance and predicted resistance based on genotype were observed. High-quality sequence data were obtained from one smear-positive culture-negative case. This study demonstrated for the first time the successful and accurate sequencing of M. tuberculosis genomes directly from uncultured sputa. Identification of known resistance mutations within a week of sample receipt offers the prospect for personalized rather than empirical treatment of drug-resistant tuberculosis, including the use of antimicrobial-sparing regimens, leading to improved outcomes.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures
Similar articles
-
Rapid detection of smear-negative Mycobacterium tuberculosis by PCR and sequencing for rifampin resistance with DNA extracted directly from slides.J Clin Microbiol. 2001 Jan;39(1):51-2. doi: 10.1128/JCM.39.1.51-52.2001. J Clin Microbiol. 2001. PMID: 11136747 Free PMC article.
-
[Mycobacterial tests].Kekkaku. 2008 Jan;83(1):43-59. Kekkaku. 2008. PMID: 18283915 Japanese.
-
Rapid Drug Susceptibility Testing of Drug-Resistant Mycobacterium tuberculosis Isolates Directly from Clinical Samples by Use of Amplicon Sequencing: a Proof-of-Concept Study.J Clin Microbiol. 2016 Aug;54(8):2058-67. doi: 10.1128/JCM.00535-16. Epub 2016 May 25. J Clin Microbiol. 2016. PMID: 27225403 Free PMC article.
-
Removing the bottleneck in whole genome sequencing of Mycobacterium tuberculosis for rapid drug resistance analysis: a call to action.Int J Infect Dis. 2017 Mar;56:130-135. doi: 10.1016/j.ijid.2016.11.422. Epub 2016 Dec 13. Int J Infect Dis. 2017. PMID: 27986491 Review.
-
Advances in the diagnosis of tuberculosis- Journey from smear microscopy to whole genome sequencing.Indian J Tuberc. 2020 Dec;67(4S):S61-S68. doi: 10.1016/j.ijtb.2020.09.026. Epub 2020 Oct 2. Indian J Tuberc. 2020. PMID: 33308673 Review.
Cited by
-
Correlation of drug resistance with single nucleotide variations through genome analysis and experimental validation in a multi-drug resistant clinical isolate of M. tuberculosis.BMC Microbiol. 2020 Jul 25;20(1):223. doi: 10.1186/s12866-020-01912-6. BMC Microbiol. 2020. PMID: 32711461 Free PMC article.
-
Evaluating the clinical impact of routine whole genome sequencing in tuberculosis treatment decisions and the issue of isoniazid mono-resistance.BMC Infect Dis. 2022 Apr 7;22(1):349. doi: 10.1186/s12879-022-07329-y. BMC Infect Dis. 2022. PMID: 35392842 Free PMC article.
-
Molecular Capture of Mycobacterium tuberculosis Genomes Directly from Clinical Samples: A Potential Backup Approach for Epidemiological and Drug Susceptibility Inferences.Int J Mol Sci. 2023 Feb 2;24(3):2912. doi: 10.3390/ijms24032912. Int J Mol Sci. 2023. PMID: 36769230 Free PMC article.
-
Culture-Free Phylogenetic Analysis of Legionella pneumophila Using Targeted CRISPR/Cas9 Next-Generation Sequencing.Microbiol Spectr. 2022 Aug 31;10(4):e0035922. doi: 10.1128/spectrum.00359-22. Epub 2022 Jul 11. Microbiol Spectr. 2022. PMID: 35862996 Free PMC article.
-
Validation of Novel Mycobacterium tuberculosis Isoniazid Resistance Mutations Not Detectable by Common Molecular Tests.Antimicrob Agents Chemother. 2018 Sep 24;62(10):e00974-18. doi: 10.1128/AAC.00974-18. Print 2018 Oct. Antimicrob Agents Chemother. 2018. PMID: 30082293 Free PMC article.
References
-
- Weyer K, Mirzayev F, Migliori GB, Van Gemert W, D'Ambrosio L, Zignol M, Floyd K, Centis R, Cirillo DM, Tortoli E, Gilpin C, de Dieu Iragena J, Falzon D, Raviglione M. 2013. Rapid molecular TB diagnosis: evidence, policy making and global implementation of Xpert MTB/RIF. Eur Respir J 42:252–271. doi:10.1183/09031936.00157212. - DOI - PubMed
-
- Drobniewski F, Nikolayevskyy V, Maxeiner H, Balabanova Y, Casali N, Kontsevaya I, Ignatyeva O. 2013. Rapid diagnostics of tuberculosis and drug resistance in the industrialized world: clinical and public health benefits and barriers to implementation. BMC Med 11:190. doi:10.1186/1741-7015-11-190. - DOI - PMC - PubMed
-
- Casali N, Nikolayevskyy V, Balabanova Y, Harris SR, Ignatyeva O, Kontsevaya I, Corander J, Bryant J, Parkhill J, Nejentsev S, Horstmann RD, Brown T, Drobniewski F. 2014. Evolution and transmission of drug-resistant tuberculosis in a Russian population. Nat Genet 46:279–286. doi:10.1038/ng.2878. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

