Evaluation of Unbiased Next-Generation Sequencing of RNA (RNA-seq) as a Diagnostic Method in Influenza Virus-Positive Respiratory Samples
- PMID: 25972420
- PMCID: PMC4473199
- DOI: 10.1128/JCM.02495-14
Evaluation of Unbiased Next-Generation Sequencing of RNA (RNA-seq) as a Diagnostic Method in Influenza Virus-Positive Respiratory Samples
Abstract
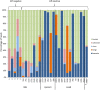
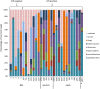
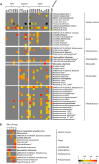
Unbiased nontargeted metagenomic RNA sequencing (UMERS) has the advantage to detect known as well as unknown pathogens and, thus, can significantly improve the detection of viral, bacterial, parasitic, and fungal sequences in public health settings. In particular, conventional diagnostic methods successfully identify the putative pathogenic agent in only 30% to 40% of respiratory specimens from patients with acute respiratory illness. Here, we applied UMERS to 24 diagnostic respiratory specimens (bronchoalveolar lavage [BAL] fluid, sputum samples, and a swab) from patients with seasonal influenza infection and 5 BAL fluid samples from patients with pneumonia that tested negative for influenza to validate RNA sequencing as an unbiased diagnostic tool in comparison to conventional diagnostic methods. In addition to our comparison to PCR, we evaluated the potential to retrieve comprehensive influenza virus genomic information and the capability to detect known superinfecting pathogens. Compared to quantitative real-time PCR for influenza viral sequences, UMERS detected influenza viral sequences in 18 of 24 samples. Complete influenza virus genomes could be assembled from 8 samples. Furthermore, in 3 of 24 influenza-positive samples, additional viral pathogens could be detected, and 2 of 24 samples showed a significantly increased abundance of individual bacterial species known to cause superinfections during an influenza virus infection. Thus, analysis of respiratory samples from known or suspected influenza patients by UMERS provides valuable information that is relevant for clinical investigation.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures


References
-
- Dawood FS, Iuliano AD, Reed C, Meltzer MI, Shay DK, Cheng PY, Bandaranayake D, Breiman RF, Brooks WA, Buchy P, Feikin DR, Fowler KB, Gordon A, Hien NT, Horby P, Huang QS, Katz MA, Krishnan A, Lal R, Montgomery JM, Molbak K, Pebody R, Presanis AM, Razuri H, Steens A, Tinoco YO, Wallinga J, Yu H, Vong S, Bresee J, Widdowson MA. 2012. Estimated global mortality associated with the first 12 months of 2009 pandemic influenza A H1N1 virus circulation: a modelling study. Lancet Infect Dis 12:687–695. doi:10.1016/S1473-3099(12)70121-4. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

