Comparison of Inoculation with the InoqulA and WASP Automated Systems with Manual Inoculation
- PMID: 25972424
- PMCID: PMC4473203
- DOI: 10.1128/JCM.03076-14
Comparison of Inoculation with the InoqulA and WASP Automated Systems with Manual Inoculation
Abstract
The quality of sample inoculation is critical for achieving an optimal yield of discrete colonies in both monomicrobial and polymicrobial samples to perform identification and antibiotic susceptibility testing. Consequently, we compared the performance between the InoqulA (BD Kiestra), the WASP (Copan), and manual inoculation methods. Defined mono- and polymicrobial samples of 4 bacterial species and cloudy urine specimens were inoculated on chromogenic agar by the InoqulA, the WASP, and manual methods. Images taken with ImagA (BD Kiestra) were analyzed with the VisionLab version 3.43 image analysis software to assess the quality of growth and to prevent subjective interpretation of the data. A 3- to 10-fold higher yield of discrete colonies was observed following automated inoculation with both the InoqulA and WASP systems than that with manual inoculation. The difference in performance between automated and manual inoculation was mainly observed at concentrations of >10(6) bacteria/ml. Inoculation with the InoqulA system allowed us to obtain significantly more discrete colonies than the WASP system at concentrations of >10(7) bacteria/ml. However, the level of difference observed was bacterial species dependent. Discrete colonies of bacteria present in 100- to 1,000-fold lower concentrations than the most concentrated populations in defined polymicrobial samples were not reproducibly recovered, even with the automated systems. The analysis of cloudy urine specimens showed that InoqulA inoculation provided a statistically significantly higher number of discrete colonies than that with WASP and manual inoculation. Consequently, the automated InoqulA inoculation greatly decreased the requirement for bacterial subculture and thus resulted in a significant reduction in the time to results, laboratory workload, and laboratory costs.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures


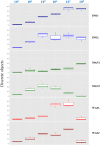
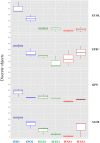



References
-
- Mischnik A, Mieth M, Busch CJ, Hofer S, Zimmermann S. 2012. First evaluation of automated specimen inoculation for wound swab samples by use of the Previ Isola system compared to manual inoculation in a routine laboratory: finding a cost-effective and accurate approach. J Clin Microbiol 50:2732–2736. doi:10.1128/JCM.05501-11. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

