Hepatitis B Virus-Infected HepG2hNTCP Cells Serve as a Novel Immunological Tool To Analyze the Antiviral Efficacy of CD8+ T Cells In Vitro
- PMID: 25972537
- PMCID: PMC4473563
- DOI: 10.1128/JVI.00605-15
Hepatitis B Virus-Infected HepG2hNTCP Cells Serve as a Novel Immunological Tool To Analyze the Antiviral Efficacy of CD8+ T Cells In Vitro
Abstract
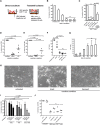
CD8(+) T cells are the main effector lymphocytes in the control of hepatitis B virus (HBV) infection. However, limitations of model systems, such as low infection rates, restrict mechanistic studies of HBV-specific CD8(+) T cells. Here, we established a novel immunological cell culture model based on HBV-infected HepG2(hNTCP) cells that endogenously processed viral antigens and presented them to HBV-specific CD8(+) T cells. This induced cytolytic and noncytolytic CD8(+) T-cell effector functions and reduction of viral loads.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

