The role of long non-coding RNAs in genome formatting and expression
- PMID: 25972893
- PMCID: PMC4413816
- DOI: 10.3389/fgene.2015.00165
The role of long non-coding RNAs in genome formatting and expression
Abstract
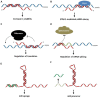
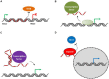
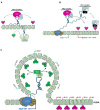
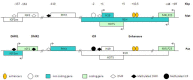
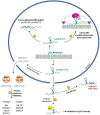
Long non-coding RNAs (lncRNAs) are transcripts without protein-coding potential but having a pivotal role in numerous biological functions. Long non-coding RNAs act as regulators at different levels of gene expression including chromatin organization, transcriptional regulation, and post-transcriptional control. Misregulation of lncRNAs expression has been found to be associated to cancer and other human disorders. Here, we review the different types of lncRNAs, their mechanisms of action on genome formatting and expression and emphasized on the multifaceted action of the H19 lncRNA.
Keywords: H19; chromatin organization; lncRNAs; post-transcriptional control; transcriptional regulation.
Figures
References
-
- Adriaenssens E., Dumont L., Lottin S., Bolle D., Leprêtre A., Delobelle A., et al. (1998). H19 overexpression in breast adenocarcinoma stromal cells is associated with tumor values and steroid receptor status but independent of p53 and Ki-67 expression. Am. J. Pathol. 153 1597–1607 10.1016/S0002-9440(10)65748-3 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

