Under-detection of endospore-forming Firmicutes in metagenomic data
- PMID: 25973144
- PMCID: PMC4427659
- DOI: 10.1016/j.csbj.2015.04.002
Under-detection of endospore-forming Firmicutes in metagenomic data
Abstract
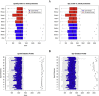
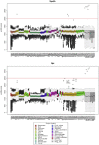
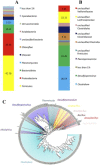
Microbial diversity studies based on metagenomic sequencing have greatly enhanced our knowledge of the microbial world. However, one caveat is the fact that not all microorganisms are equally well detected, questioning the universality of this approach. Firmicutes are known to be a dominant bacterial group. Several Firmicutes species are endospore formers and this property makes them hardy in potentially harsh conditions, and thus likely to be present in a wide variety of environments, even as residents and not functional players. While metagenomic libraries can be expected to contain endospore formers, endospores are known to be resilient to many traditional methods of DNA isolation and thus potentially undetectable. In this study we evaluated the representation of endospore-forming Firmicutes in 73 published metagenomic datasets using two molecular markers unique to this bacterial group (spo0A and gpr). Both markers were notably absent in well-known habitats of Firmicutes such as soil, with spo0A found only in three mammalian gut microbiomes. A tailored DNA extraction method resulted in the detection of a large diversity of endospore-formers in amplicon sequencing of the 16S rRNA and spo0A genes. However, shotgun classification was still poor with only a minor fraction of the community assigned to Firmicutes. Thus, removing a specific bias in a molecular workflow improves detection in amplicon sequencing, but it was insufficient to overcome the limitations for detecting endospore-forming Firmicutes in whole-genome metagenomics. In conclusion, this study highlights the importance of understanding the specific methodological biases that can contribute to improve the universality of metagenomic approaches.
Keywords: Endospores; Metagenomics; Profile analysis; gpr; spo0A.
Figures
References
-
- Suenaga H. Targeted metagenomics: a high-resolution metagenomics approach for specific gene clusters in complex microbial communities. Environ Microbiol. 2012;14:13–22. - PubMed
-
- Xu J. Microbial ecology in the age of genomics and metagenomics: concepts, tools, and recent advances. Mol Ecol. 2006;15:1713–1731. - PubMed
-
- Béjà O., Suzuki M.T., Heidelberg J.F., Nelson W.C., Preston C.M., Hamada T. Unsuspected diversity among marine aerobic anoxygenic phototrophs. Nature. 2002;415:630–633. - PubMed
-
- Béjà O., Aravind L., Koonin E.V., Suzuki M.T., Hadd A., Nguyen L.P. Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science. 2000;289:1902–1906. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
