Identification of miRNAs contributing to neuroblastoma chemoresistance
- PMID: 25973145
- PMCID: PMC4427660
- DOI: 10.1016/j.csbj.2015.04.003
Identification of miRNAs contributing to neuroblastoma chemoresistance
Abstract
Background: The emergence of the role of microRNAs (miRNAs) in exacerbating drug resistance of tumours is recently being highlighted as a crucial research field for future clinical management of drug resistant tumours. The purpose of this study was to identify dys-regulations in expression of individual and/or networks of miRNAs that may have direct effect on neuroblastoma (NB) drug resistance.
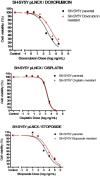
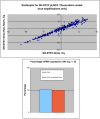
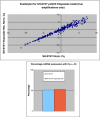
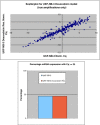
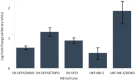
Methods: Individual subcultures of chemosensitive SH-SY5Y and UKF-NB-3 cells were rendered chemoresistant to doxorubicin (SH-SY5Y, UKF-NB-3) or etoposide (SH-SY5Y). In each validated chemoresistance model, the parental and subcultured cell lines were analysed for miRNA expression profiling, using a high-throughput quantitative polymerase chain reaction (RT-qPCR) miRNA profiling platform for a total of 668 miRNAs.
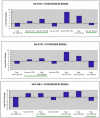
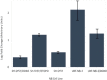
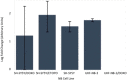
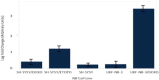
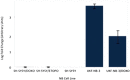
Results: A unique expression signature of miRNAs was found to be differentially expressed (higher than 2-fold change) within all three NB chemoresistance models. Four miRNAs were upregulated in the subcultured chemoresistant cell line. Three miRNAs were found to be downregulated in the chemoresistant cell lines for all models.
Conclusions: Based on the initial miRNA findings, this study elucidates the dys-regulation of four miRNAs in three separate NB chemoresistant cell line models, spanning two cell lines (SH-SY5Y and UKF-NB-3) and two chemotherapeutic agents (doxorubicin and etoposide). These miRNAs may thus be possibly linked to chemoresistance induction in NB. Such miRNAs are good candidates to be novel drug targets for future miRNA based therapies against aggressive tumours that are not responding to conventional chemotherapy.
Keywords: Chemoresistance; Drug; Neuroblastoma; Resistance; miRNA.
Figures





Similar articles
-
Characterization of drug-resistant neuroblastoma cell lines by comparative genomic hybridization.Neoplasma. 2005;52(5):415-9. Neoplasma. 2005. PMID: 16151587
-
Chemoresistance induces enhanced adhesion and transendothelial penetration of neuroblastoma cells by down-regulating NCAM surface expression.BMC Cancer. 2006 Dec 21;6:294. doi: 10.1186/1471-2407-6-294. BMC Cancer. 2006. PMID: 17181871 Free PMC article.
-
Antitumor activity of the selective MDM2 antagonist nutlin-3 against chemoresistant neuroblastoma with wild-type p53.J Natl Cancer Inst. 2009 Nov 18;101(22):1562-74. doi: 10.1093/jnci/djp355. Epub 2009 Nov 10. J Natl Cancer Inst. 2009. PMID: 19903807
-
The roles of microRNAs in neuroblastoma.World J Pediatr. 2014 Feb;10(1):10-6. doi: 10.1007/s12519-014-0448-2. Epub 2014 Jan 25. World J Pediatr. 2014. PMID: 24464658 Review.
-
Role of microRNAs in chemoresistance.Ann Transl Med. 2015 Dec;3(21):332. doi: 10.3978/j.issn.2305-5839.2015.11.32. Ann Transl Med. 2015. PMID: 26734642 Free PMC article. Review.
Cited by
-
Determination of absolute expression profiles using multiplexed miRNA analysis.PLoS One. 2017 Jul 13;12(7):e0180988. doi: 10.1371/journal.pone.0180988. eCollection 2017. PLoS One. 2017. PMID: 28704432 Free PMC article.
-
MicroRNAs in neural crest development and neurocristopathies.Biochem Soc Trans. 2022 Apr 29;50(2):965-974. doi: 10.1042/BST20210828. Biochem Soc Trans. 2022. PMID: 35383827 Free PMC article. Review.
-
miR-149 inhibits cell proliferation and enhances chemosensitivity by targeting CDC42 and BCL2 in neuroblastoma.Cancer Cell Int. 2019 Dec 27;19:357. doi: 10.1186/s12935-019-1082-9. eCollection 2019. Cancer Cell Int. 2019. PMID: 31889909 Free PMC article.
-
Hsa-miR-323a-3p functions as a tumor suppressor and targets STAT3 in neuroblastoma cells.Front Pediatr. 2023 Mar 24;11:1098999. doi: 10.3389/fped.2023.1098999. eCollection 2023. Front Pediatr. 2023. PMID: 37033189 Free PMC article.
-
Comprehensive Characterization of Circular RNAs in Neuroblastoma Cell Lines.Technol Cancer Res Treat. 2020 Jan-Dec;19:1533033820957622. doi: 10.1177/1533033820957622. Technol Cancer Res Treat. 2020. PMID: 33000697 Free PMC article.
References
-
- Izbicka E., Izbicki T. Therapeutic strategies for the treatment of neuroblastoma. Curr Opin Investig Drugs. Dec 2005;6(12):1200–1214. - PubMed
-
- American Cancer Society. What are the most common types of childhood cancers? [Internet]. 2010 [cited 2010 Nov 28]. Available from: http://www.cancer.org/Cancer/CancerinChildren/DetailedGuide/cancer-in-ch...
-
- Kushner B.H., Cheung N.-K.V. Neuroblastoma—from genetic profiles to clinical challenge. N Engl J Med. Nov 24 2005;353(21):2215–2217. - PubMed
-
- Grau E., Oltra S., Orellana C., Hernández-Martí M., Castel V., Martínez F. There is no evidence that the SDHB gene is involved in neuroblastoma development. Oncol Res. 2005;15(7-8):393–398. - PubMed
-
- Magro G., Grasso S. The glial cell in the ontogenesis of the human peripheral sympathetic nervous system and in neuroblastoma. Pathologica. Oct 2001;93(5):505–516. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
