Comparison of Exome and Genome Sequencing Technologies for the Complete Capture of Protein-Coding Regions
- PMID: 25973577
- PMCID: PMC4755152
- DOI: 10.1002/humu.22813
Comparison of Exome and Genome Sequencing Technologies for the Complete Capture of Protein-Coding Regions
Abstract
For next-generation sequencing technologies, sufficient base-pair coverage is the foremost requirement for the reliable detection of genomic variants. We investigated whether whole-genome sequencing (WGS) platforms offer improved coverage of coding regions compared with whole-exome sequencing (WES) platforms, and compared single-base coverage for a large set of exome and genome samples. We find that WES platforms have improved considerably in the last years, but at comparable sequencing depth, WGS outperforms WES in terms of covered coding regions. At higher sequencing depth (95x-160x), WES successfully captures 95% of the coding regions with a minimal coverage of 20x, compared with 98% for WGS at 87-fold coverage. Three different assessments of sequence coverage bias showed consistent biases for WES but not for WGS. We found no clear differences for the technologies concerning their ability to achieve complete coverage of 2,759 clinically relevant genes. We show that WES performs comparable to WGS in terms of covered bases if sequenced at two to three times higher coverage. This does, however, go at the cost of substantially more sequencing biases in WES approaches. Our findings will guide laboratories to make an informed decision on which sequencing platform and coverage to choose.
Keywords: coverage; exome sequencing; genome sequencing.
© 2015 WILEY PERIODICALS, INC.
Figures

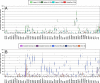
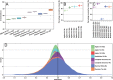
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

