XBP1 mitigates aminoglycoside-induced endoplasmic reticulum stress and neuronal cell death
- PMID: 25973683
- PMCID: PMC4669688
- DOI: 10.1038/cddis.2015.108
XBP1 mitigates aminoglycoside-induced endoplasmic reticulum stress and neuronal cell death
Abstract
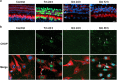
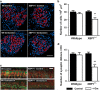
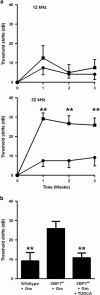
Here we study links between aminoglycoside-induced mistranslation, protein misfolding and neuropathy. We demonstrate that aminoglycosides induce misreading in mammalian cells and assess endoplasmic reticulum (ER) stress and unfolded protein response (UPR) pathways. Genome-wide transcriptome and proteome analyses revealed upregulation of genes related to protein folding and degradation. Quantitative PCR confirmed induction of UPR markers including C/EBP homologous protein, glucose-regulated protein 94, binding immunoglobulin protein and X-box binding protein-1 (XBP1) mRNA splicing, which is crucial for UPR activation. We studied the effect of a compromised UPR on aminoglycoside ototoxicity in haploinsufficient XBP1 (XBP1(+/-)) mice. Intra-tympanic aminoglycoside treatment caused high-frequency hearing loss in XBP1(+/-) mice but not in wild-type littermates. Densities of spiral ganglion cells and synaptic ribbons were decreased in gentamicin-treated XBP1(+/-) mice, while sensory cells were preserved. Co-injection of the chemical chaperone tauroursodeoxycholic acid attenuated hearing loss. These results suggest that aminoglycoside-induced ER stress and cell death in spiral ganglion neurons is mitigated by XBP1, masking aminoglycoside neurotoxicity at the organismal level.
Figures



Comment in
-
Stressing out the ER in aminoglycoside-induced hearing loss.Cell Death Dis. 2015 May 14;6(5):e1762. doi: 10.1038/cddis.2015.133. Cell Death Dis. 2015. PMID: 25973682 Free PMC article. No abstract available.
References
-
- Prostko CR, Brostrom MA, Malara EM, Brostrom CO. Phosphorylation of eukaryotic initiation factor (eIF) 2 alpha and inhibition of eIF-2B in GH3 pituitary cells by perturbants of early protein processing that induce GRP78. J Biol Chem 1992; 267: 16751–16754. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

